Author(s): Michele D’Orazio, Riccardo Reale, Adele De Ninno, Maria A. Brighetti, Arianna Mencattini, Luca Businaro, Eugenio Martinelli, Paolo Bisegna, Alessandro Travaglini, and Federica Caselli
Palynology is an interdisciplinary science that deals with the study of pollen and fungal spores and finds application in many fields. As an example, in agriculture, control over pollen quality along the pollen supply chain is a key success factor to ensure high quality seed and yield. On the other hand, aerobiological monitoring represents a critical tool to improve the quality of life in an urban environment.
Traditionally, the study of pollen and fungal spores takes place through microscopic analysis of samples by specialized operators and usually involves labelling through a colorimetric or fluorescence stain. The procedure requires long time and, despite being regulated by standardized methodologies, it is subject to errors. Therefore, there is a pressing need for accurate, label-free and automated analysis of pollen grains.
To tackle this need, we propose a novel multimodal approach that combines electrical sensing and optical imaging. The operating principle is as follows: pollen grains from a mixture of taxa flow one-by-one through a microfluidic chip; the variation in channel impedance due to the passage of individual grains is recorded with an impedance spectroscope, while acquiring a high-speed optical video. Electrical signals and synchronized optical images are processed by two independent machine learning-based classifiers, providing the electrical and optical probability of belonging to the different taxa, respectively. These probabilities are then combined by applying a consensus paradigm to obtain the classification outcome.
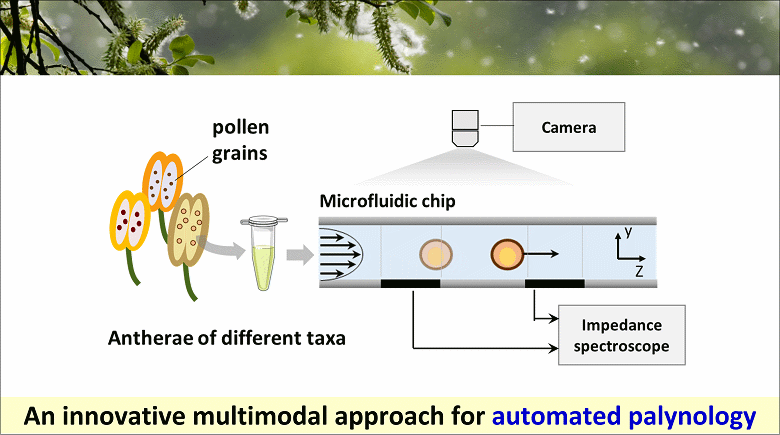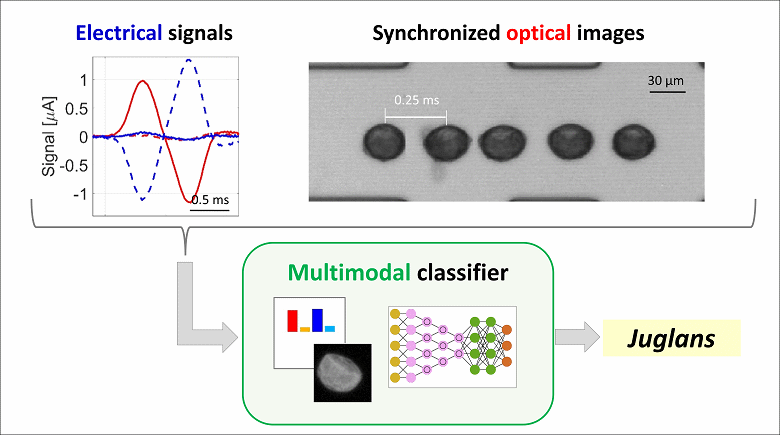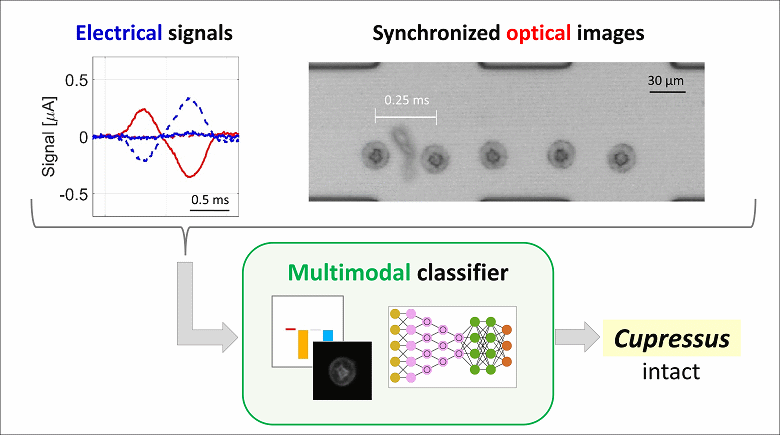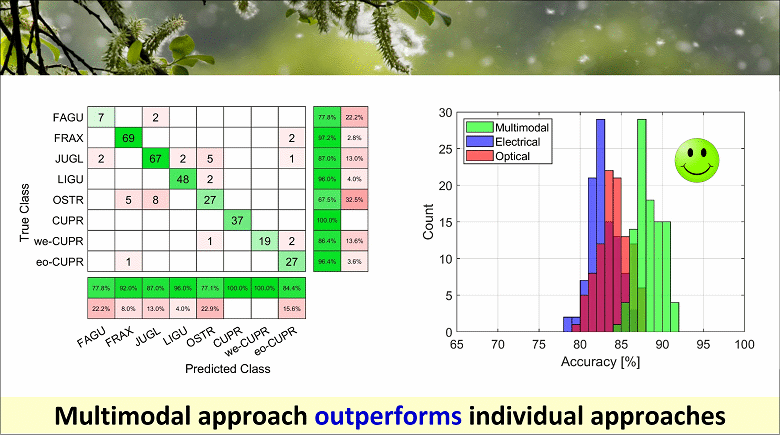
As demonstrated in a proof-of-concept classification experiment, this multimodal approach provides better accuracy with respect to the analysis based on electrical or optical features alone. Moreover, using the electrical signals as pointers to image frames significantly reduces the burden of image processing. In addition, the proposed methodology can be extended to other fields, such as diagnostics and cell therapy, for label-free identification of cell populations in heterogeneous samples.

