Generalizable Pancreas Segmentation via a Dual Self-Supervised Learning Framework
Qian, Xiaohua; Li, Jun; Zhu, Hongzhang; Chen, Tao
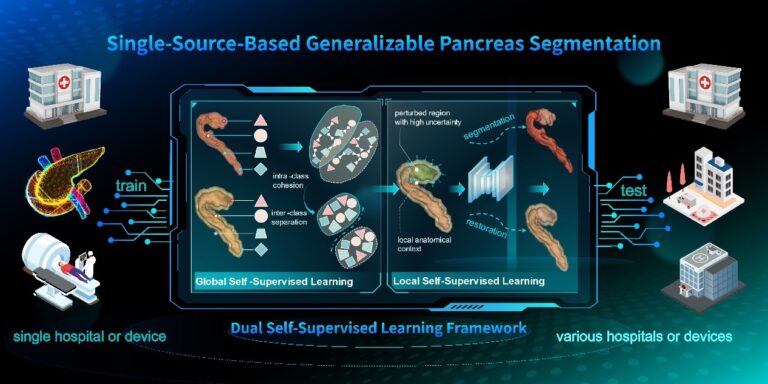
Recently, numerous pancreas segmentation methods have achieved promising performance on local single-source datasets. However, these methods don’t adequately account for generalizability issues, and hence typically show limited performance and low stability on test data from other sources. Considering the limited availability of distinct data sources, we seek to improve the generalization performance of a pancreas segmentation model trained with a single-source dataset, i.e., the single-source generalization task. In particular, we propose a dual self-supervised learning model that incorporates both global and local anatomical contexts. Our model aims to fully exploit the anatomical features of the intra-pancreatic and extra-pancreatic regions, and hence enhance the characterization of the high-uncertainty regions for more robust generalization. Specifically, we first construct a global-feature contrastive self-supervised learning module that is guided by the pancreatic spatial structure. This module obtains complete and consistent pancreatic features through promoting intra-class cohesion, and also extracts more discriminative features for differentiating between pancreatic and non-pancreatic tissues through maximizing inter-class separation. It mitigates the influence of surrounding tissue on the segmentation outcomes in high-uncertainty regions. Subsequently, a local-image-restoration self-supervised learning module is introduced to further enhance the characterization of the high-uncertainty regions. In this module, informative anatomical contexts are actually learned to recover randomly-corrupted appearance patterns in those regions. The effectiveness of our method is demonstrated with state-of-the-art performance and comprehensive ablation analysis on three pancreas datasets (467 cases). The results demonstrate a great potential in providing a stable support for the diagnosis and treatment of pancreatic diseases.