IEEE-EMBS International Conference on Biomedical and Health Informatics (BHI) 2024
10 – 13 November 2024 Houston, TX USA
Discovering consensus regions for interpretable identification of RNA N6-methyladenosine modification sites via graph contrastive clustering
G. Li, B. Zhao, X. Su, Y. Yang, P. Hu, X. Zhou, L. Hu
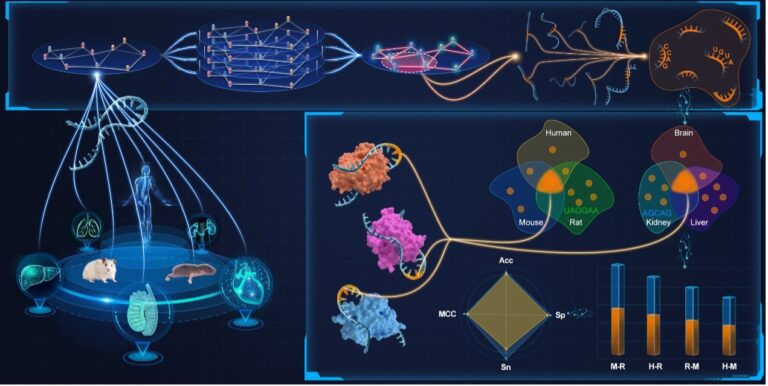
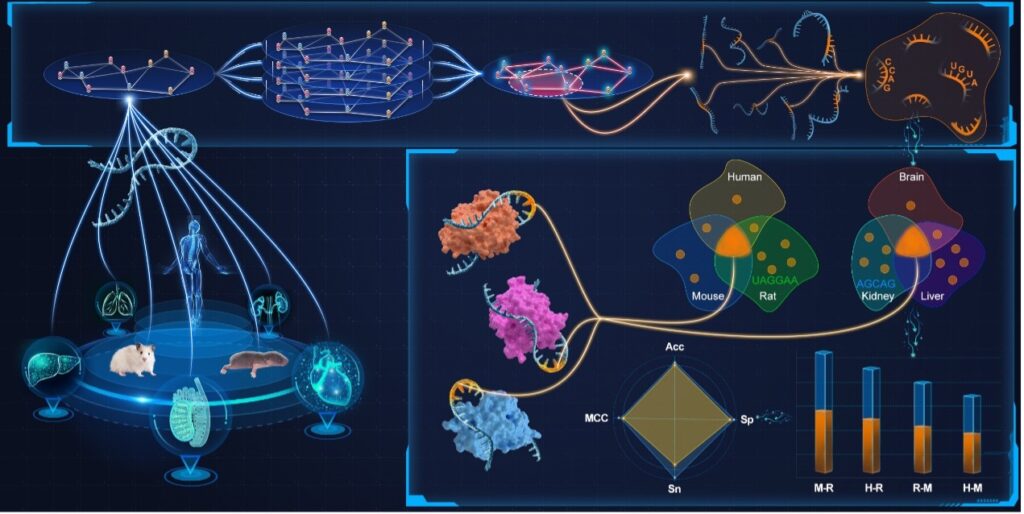
As a pivotal post-transcriptional modification of RNA, N6-methyladenosine (m6A) has a substantial influence on gene expression modulation and cellular fate determination. Although a variety of computational models have been developed to accurately identify potential m6A modification sites, few of them are capable of interpreting the identification process with insights gained from consensus knowledge. To overcome this problem, we propose a deep learning model, namely M6A-DCR, by discovering consensus regions for interpretable identification of m6A modification sites. In particular, M6A-DCR first constructs an instance graph for each RNA sequence by integrating specific positions and types of nucleotides. The discovery of consensus regions is then formulated as a graph clustering problem in light of aggregating all instance graphs. After that, M6A-DCR adopts a motif-aware graph reconstruction optimization process to learn high-quality embeddings of input RNA sequences, thus achieving the identification of m6A modification sites in an end-to-end manner. Experimental results demonstrate the superior performance of M6A-DCR by comparing it with several state-of-the-art identification models. The consideration of consensus regions empowers our model to make interpretable predictions at the motif level. The analysis of cross validation through different species and tissues further verifies the consistency between the identification results of M6A-DCR and the evolutionary relationships among species.
Sensor Informatics
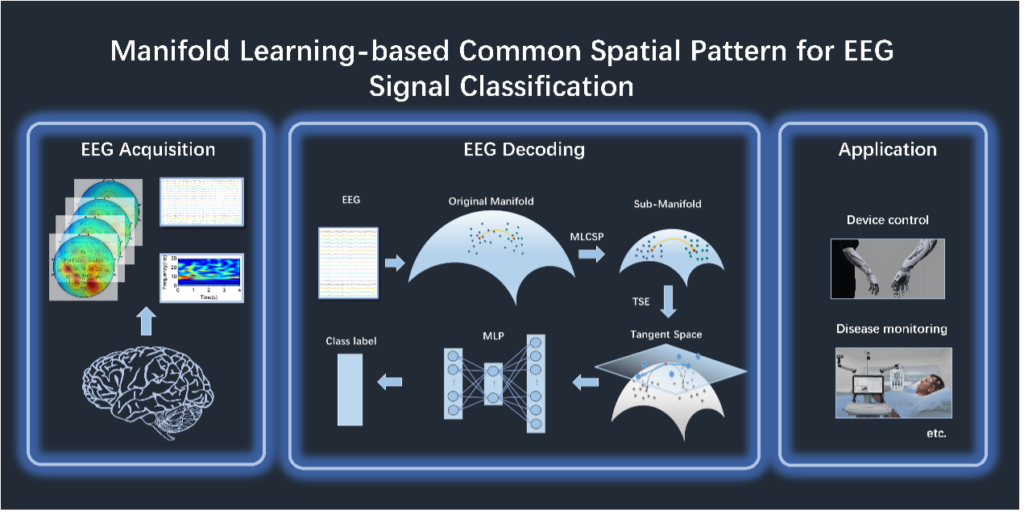
Manifold Learning-based Common Spatial Pattern for EEG Signal Classification
Ma, Ting; Cai, Guoqing; Zhang, Fenghui; Yang, Bolun; Huang, Shoulin
Multi-view Cross-Fusion Transformer Based on Kinetic Features for Non-invasive Blood Glucose Measurement using PPG Signal
Zhang, Yuan; Chen, Shisen; Qin, Fen; Ma, Xuesheng; Wei, Jie; Zhang, Yuanting; Jovanov, Emil
Elimination of Random Mixed Noise in ECG using Convolutional Denoising Autoencoder with Transformer Encoder
Wei, Shoushui; Chen, Meng; Li, Yongjian; Zhang, Liting; Liu, Lei; Han, Baokun; Shi, Wenzhuo
MD-CardioNet: A Multi-Dimensional Deep Neural Network for Cardiovascular Disease Diagnosis from Electrocardiogram
Tahmid, Md; Kader, Muhammad Ehsanul; Mahmud, Tanvir; Fattah, Shaikh Anowarul
ECG Feature Importance Rankings: Cardiologists vs. Algorithms
Mehari, Temesgen; Sundar, Ashish; Bosnjakovic, Alen; Harris, Peter; Williams, Steven E.; Loewe, Axel; Doessel, Olaf; Nagel, Claudia; Strodthoff, Nils; Aston, Philip
Generative Listener EEG for Speech Emotion Recognition Using Generative Adversarial Networks with Compressed Sensing
Chang, Jiang; Zhang, Zhixin; Wang, Zelin; Li, Jiacheng; Meng, Linsheng; Lin, Pan
Spatiotemporal Network based on GCN and BiGRU for seizure detection
Xu, Jie; Yuan, Shasha; Shang, Junliang; Wang, Juan; Yan, Kuiting; Yang, Yankai
Imaging Informatics
 Identifying biases in a multicenter MRI database for Parkinson’s disease classification: Is the disease classifier a secret site classifier?
Identifying biases in a multicenter MRI database for Parkinson’s disease classification: Is the disease classifier a secret site classifier?
Souza, Raissa; Winder, Anthony; Stanley, Emma A.M.; Vigneshwaran, Vibujithan; Camacho, Milton; Camicioli, Richard; Monchi, Oury; Wilms, Matthias; Forkert, Nils Daniel
Dense Contrastive-based Federated Learning for Dense Prediction Tasks on Medical Images
Yang, Yuning; Liu, Xiaohong; Gao, Tianrun; Xu, Xiaodong; Zhang, Ping; Wang, Guangyu
A Unified Multi-Modality Fusion Framework for Deep Spatio-Temporal-Spectral Feature Learning in Resting-State fMRI Denoising
Lim, Min-Joo; Heo, Keun-Soo; Kim, Jun-Mo; Kang, Bo-Gyeong; Lin, Weili; Zhang, Han; Shen, Dinggang; Kam, Tae-Eui
A Clinically Explainable AI-Based Grading System for Age-Related Macular Degeneration Using Optical Coherence Tomography
El-Baz, Ayman; Elsharkawy, M.; Sharafeldeen, Ahmed; Khalifa, Fahmi; Soliman, A.; Elnakib, Ahmed; Ghazal, Mohammed; Sewelam, A.; Thanos, A.; Sandhu, H. S.
Continuous Refinement-based Digital Pathology Image Assistance Scheme in Medical Decision-Making Systems
WU, Jia; Luo, Tian; Zeng, Jiachen; Gou, Fangfang
CORONet: A Cross-Sequence Joint Representation and Hypergraph Convolutional Network for Classifying Molecular Subtypes of Breast Cancer Using Incomplete DCE-MRI
Zhao, Fengjun; Xie, Xiaoyang; Wu, Lin; Su, Zhiming; Sun, Zhipeng; Cao, Xin; Hou, Yuqing; He, Xiaowei
Hybrid Masked Image Modeling for 3D Medical Image Segmentation
Wan, Liang; Xing, Zhaohu; Zhu, Lei; Yu, Lequan ; Xing, Zhiheng
A Faithful Deep Sensitivity Estimation for Accelerated Magnetic Resonance Imaging
Qu, Xiaobo; Wang, Zi; Fang, Haoming; Qian, Chen; Shi, Boxuan; Bao, Lijun; Zhu, Liuhong; Zhou, Jianjun; Wei, Wenping; Lin, Jianzhong; Guo, Di
Generalizable Polyp Segmentation via Randomized Global Illumination Augmentation
Zhang, Zuyu; Li, Yan; Shin, Byeong-Seok
TSGET: Two-Stage Global Enhanced Transformer for Automatic Radiology Report Generation
Hua, Rong; Yi, Xiulong; Fu, You; Liu, Ruiqing; Zhang, Hao
Structure-aware Registration Network for Liver DCE-CT Images
Xue, Peng; Zhang, Jingyang; Ma, Lei; Liu, Mianxin; Gu, Yuning; Huang, Jiawei; Liu, Feihong; Pan, Yongsheng; Cao, Xiaohuan; Shen, Dinggang
Three-Direction Fusion for Accurate Volumetric Liver and Tumor Segmentation
Zhan, Feng; Wang, Wenwu; Chen, Qian; Guo, Yina; He, Lidan; Wang, Lili
TNCB: Tri-net with Cross-Balanced Pseudo Supervision for Class Imbalanced Medical Image Classification
Liu, Ju; Qu, aixi; Wu, Qiang; wang, jing; Yu, LuYue; Li, Jing
CAMANet: Class Activation Map Guided Attention Network for Radiology Report Generation
Wang, Jun; Bhalerao, Abhir; Yin, Terry; See, Simon; He, Yulan
Anatomical Prior and Inter-slice Consistency for Semi-supervised Vertebral Structure Detection in 3D Ultrasound Volume
Zeng, Hong-Ye; Zhou, Kang; Ge, Song-Han; Gao, Yu-Chong; Zhao, Jianhao; Gao, Shenghua; zheng, rui
Brain Structural Connectivity Guided Vision Transformers for Identification of Functional Connectivity Characteristics in Preterm Neonates
Jiang, Xi; Mao, Wei; Chen, Yuzhong; He, Zhibin; Wang, Zifan; Xiao, Zhenxiang; Sun, Yusong; He, Liang; Zhou, Jingchao; Guo, Weitong; Ma, Chong; Zhao, Lin; Kendrick, Keith M; Zhou, Bo; Becker, Benjamin; Liu, Tianming; Zhang, Tuo
Deep Multimodal Fusion of Data with Heterogeneous Dimensionality via Projective Networks
Morano, José; Aresta, Guilherme; Grechenig, Christoph; Schmidt-Erfurth, Ursula; Bogunovic, Hrvoje
Medical Informatics
Attention Mechanisms in Clinical Text Classification: A Comparative Evaluation
Metzner, Christoph S.; Gao, Shang; Herrmannova, Drahomira; Lima-Walton, Elia; Hanson, Heidi
Predictive Modeling for Hospital Readmissions for Patients with Heart Disease: An updated review from 2012-2023
Zhu, Chengyan; Zhang, Wei; Cheng, Weihan; Fujiwara, Koichi; Evans, Richard
PVR-Vocoder: A Pathological Voice Repair Vocoder for Voice Disorders
Liu, Ganjun; Zhang, Tao; Liu, Xiaonan; Hou, Xiaohui; Fu, Dehui; Pang, Zhibo
DeepHealthNet: Adolescent Obesity Prediction System Based on a Deep Learning Framework
Jeong, Ji-Hoon; Lee, In-Gyu; Kim, Sung-Kyung; Kam, Tae-Eui; Lee, Seong-Whan; Lee, Euijong
Medicine Package Recommendation via Dual-level Interaction Aware Heterogeneous Graph
Zhu, Fanglin; Zhang, Xu; Zhang, Batuo; Xu, Yonghui; Cui, LiZhen
The ChAMP App: A Scalable mHealth Technology for Detecting Digital Phenotypes of Early Childhood Mental Health
McGinnis, Ryan; Loftness, Bryn C.; Halvorson-Phelan, Julia; OLeary, Aisling; Bradshaw, Carter; Prytherch, Shania; Berman, Isabel; Torous, John; Copeland, William; Cheney, Nick; McGinnis, Ellen
LoGo-GR: a local to global graphical reasoning framework for extracting structured information from biomedical literature
Fu, Qiming; Zhou, Xueyang; Xia, Youbing; Wang, Yunzhe; Lu, You; Chen, Yanming; Chen, Jianping
Bioinformatics
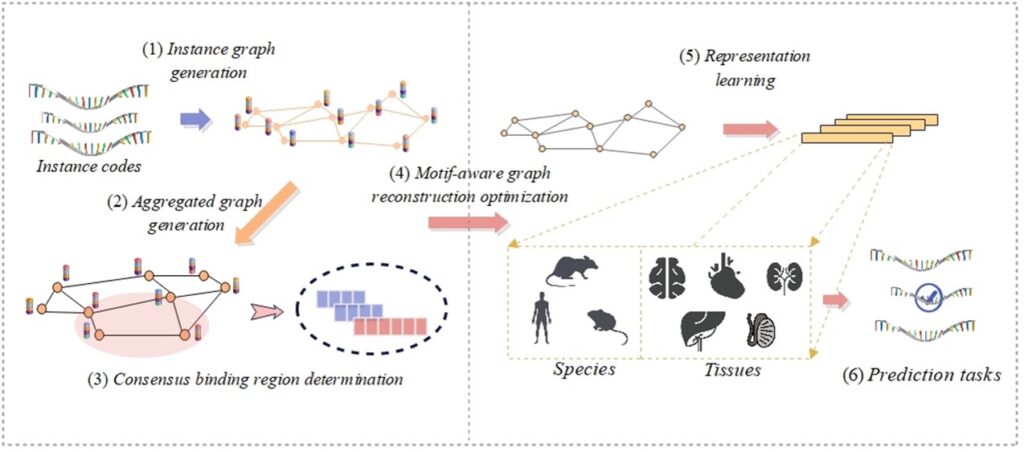 Discovering consensus regions for interpretable identification of RNA N6-methyladenosine modification sites via graph contrastive clustering
Discovering consensus regions for interpretable identification of RNA N6-methyladenosine modification sites via graph contrastive clustering
Li, Guodong; Zhao, Bowei; Su, Xiaorui; Yang, Yue; HU, Pengwei; Zhou, Xi; Hu, Lun
Network Activity Evaluation Reveals Significant Gene Regulatory Architectures during SARS-CoV-2 Viral Infection from Dynamic scRNA-seq Data
Liu, Zhi-Ping; Wang, ChuanYuan
TEPI: T_ axonomy-aware E_ mbedding and P_ seudo- I_maging for Scarcely-labeled Zero-shot Genome Classification
Aakur, Sathyanarayanan; Laguduva, Vishalini R.; Ramamurthy, Priyadharsini; Ramachandran, Akhilesh
TBCA: Prediction of transcription factor binding sites using a deep neural network with lightweight attention mechanism
Qiao, Lian; Wang, Xun; Qu, Peng; Yang, Qing
Predicting Protein Functions based on Heterogeneous Graph Attention Technique
Zhao, Yingwen; Yang, Zhihao; wang, lei; Zhang, Yin; Lin, Hongfei; Wang, Jian
Hierarchical and dynamic graph attention network for drug-disease association prediction
Huang, Shuhan; Wang, Minhui; Zheng, Xiao; Chen, Jiajia; Tang, Chang
MUMA: a multi-omics meta-learning algorithm for data interpretation and classification
Huang, Hai-Hui; Shu, Jun; Liang, Yong
Efficient Motif Discovery in Protein Sequences Using a Branch and Bound Algorithm
Neamatollahi, Peyman; Mohammadi, Rahele; Moradi, Morteza; Naghibzadeh, Mahmoud; Savadi, Abdorreza
PLANNER: a multi-scale deep language model for the origins of replication site prediction
Li, Fuyi; Wang, Cong; He, Zhijie; Jia, Runchang; Pan, Shirui ; JM, Lachlan Coin; Song, Jiangning
Modeling and AI Informatics
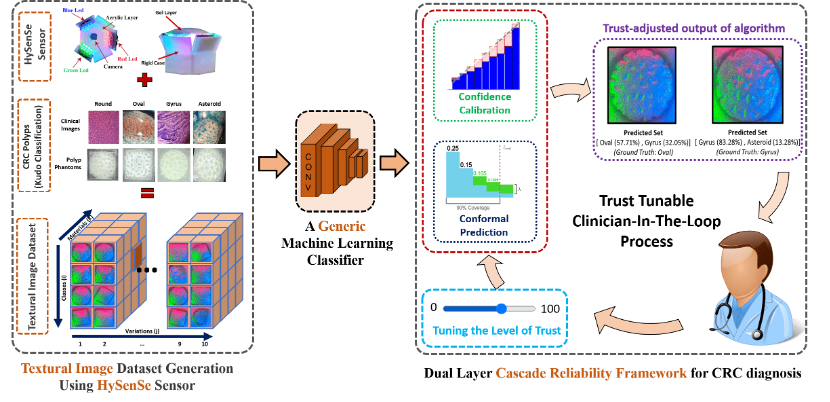 A Novel Dual Layer Cascade Reliability Framework for an Informed and Intuitive Clinician-AI Interaction In Diagnosis of Colorectal Cancer Polyps
A Novel Dual Layer Cascade Reliability Framework for an Informed and Intuitive Clinician-AI Interaction In Diagnosis of Colorectal Cancer Polyps
Alambeigi, Farshid; Kapuria, Siddhartha; Minot, Patrick; Kapusta, Ariel; Ikoma, Naruhiko
RClaNet: An explainable Alzheimer’s disease diagnosis framework by joint deformable registration and classification
WU, Liang; Hu, Shunbo; Wang, Duanwei; Liu, Changchun; Wang, Li
Data augmentation for Human Activity Recognition with Generative Adversarial Networks
Lupión, Marcos; Cruciani, Federico ; Cleland, Ian; Nugent, Chris; Ortigosa, Pilar M.
Insights of Machine Learning into Medical Decision Making Systems: From Research to Practice
An overview of data integration in neuroscience with focus on Alzheimer’s Disease
Turrisi, Rosanna; Squillario, Margherita; Abate, Giulia; Uberti, Daniela; Barla, Annalisa
Identification of congenital valvular murmurs in young patients using deep learning-based attention transformers and phonocardiograms
Alkhodari, Mohanad; Hadjileontiadis, Leontios; Khandoker, Ahsan
Convolutional Attention Based Multimodal Network for Depression Detection on Social Media and Its Impact During Pandemic
Zogan, Hamad; Razzak, Imran; Jameel, Shoaib; Xu, Guandong
Knowledge-enhanced Graph Topic Transformer for Explainable Biomedical Text Summarization
Xie, Qianqian; Tiwari, Prayag; Ananiadou, Sophia
Analysis of a Deep Learning Model for 12-Lead ECG Classification Reveals Learned Features Similar to Diagnostic Criteria
Bender, Theresa; Beinecke, Jacqueline M.; Krefting, Dagmar; Müller, Carolin; Dathe, Henning; Seidler, Tim; Spicher, Nicolai; Hauschild, Anne-Christin
Explainable Early Prediction of Gestational Diabetes Biomarkers by Combining Medical Background and Wearable Devices: A Pilot Study in South Africa
Kolozali, Sefki; L, Sara White; Norris, Shane; Fasli, Maria; Heerden, Alastair van
Few-shot class-incremental learning for Medical time series classification
SUN, Le; Zhang, Mingyang; Wang, Benyou; Tiwari, Prayag
Advanced Machine Learning and Artificial Intelligence Tools for Computational Biology
Exploiting Hierarchical Interactions for Protein Surface Learning
Lin, Yiqun; Pan, Liang; Li, Yi; Liu, Ziwei; Li, Xiaomeng
EnDL-HemoLyt: Ensemble deep learning-based tool for identifying therapeutic peptides with low hemolytic activity
Sharma, Ritesh; Shrivastava, Sameer; Singh, Sanjay Kumar; Kumar, Abhinav; Singh, Amit Kumar; Saxena, Sonal
Employing Feature Selection Algorithms to Determine the Immune State of Mice Model of Rheumatoid Arthritis
Talitckii, Aleksandr; Mangal, Joslyn L.; Colbert, Brendon K.; Acharya, Abhinav P.; Peet, Matthew M.
Hybrid Bayesian Optimization-based Graphical Discovery for Methylation Sites Prediction
Gu, Ling-Yan; Chen, Tingbo; Li, Jianqiang; Huang, Yu-An; Du, Zhihua; Leung, Victor; Chen, Jie
K-PathVQA: Knowledge-Aware Multimodal Representation for Pathology Visual Question Answering
Naseem, Usman; Khushi, Matloob ; Dunn, Adam G.; Kim, Jinman
Prediction of LncRNA-Protein Interactions Based on Kernel Combinations and Graph Convolutional Networks
Shen, Cong; Mao, Dongdong; Tang, Jijun; Liao, Zhijun; Chen, S.Y.
Artificial intelligence-based model for predicting the minimum inhibitory concentration of antibacterial peptides against ESKAPEE pathogens
Sharma, Ritesh; Shrivastava, Sameer; Singh, Sanjay Kumar; Kumar, Abhinav; Singh, Amit Kumar; Saxena, Sonal
Compound Scaling Encoder-Decoder (CoSED) Network for Diabetic Retinopathy Related Bio-marker Detection
Yi, Dewei; Baltov, Petar; Hua, Yining; Philip, Sam; Sharma, Pradip