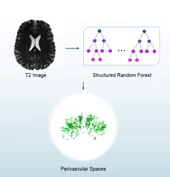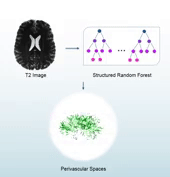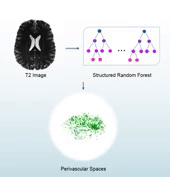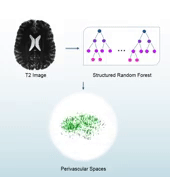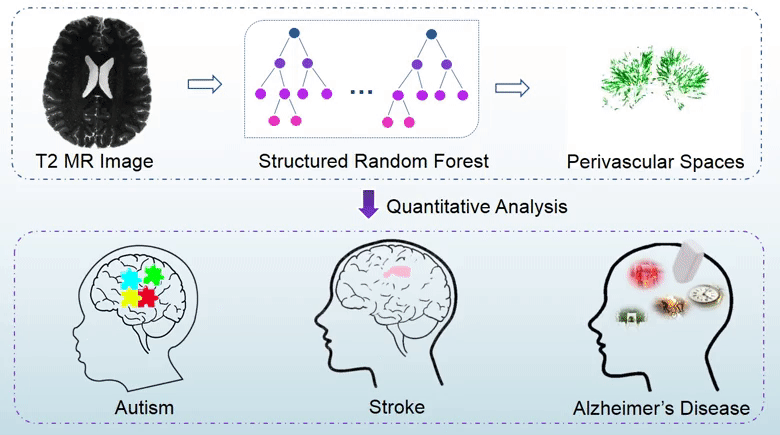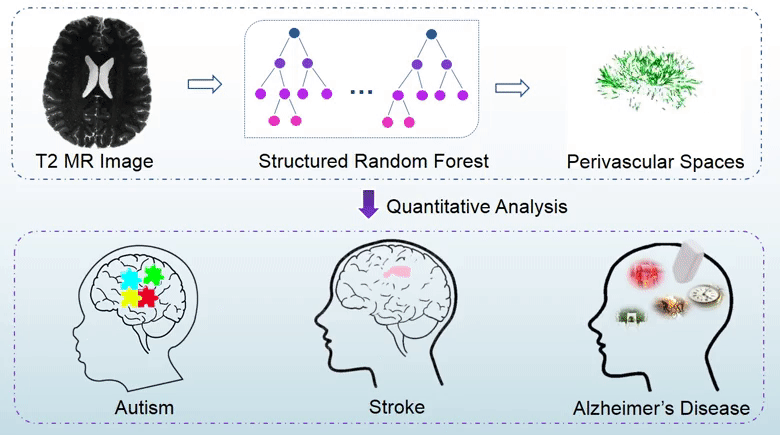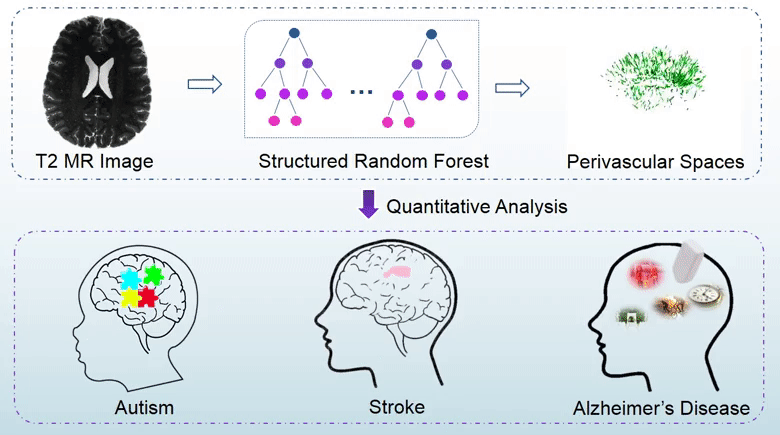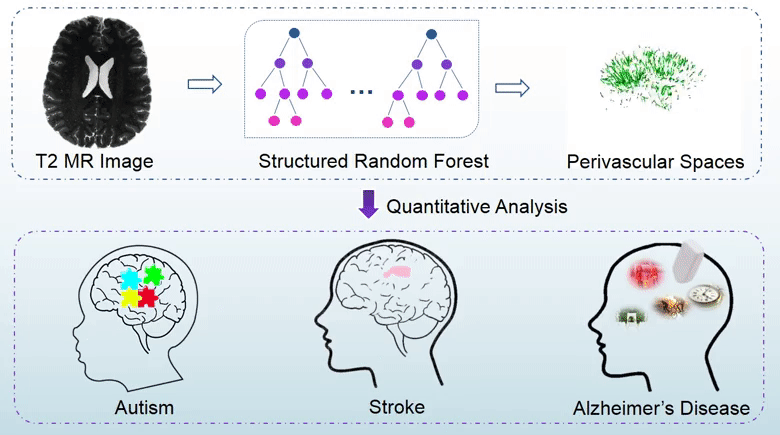
Perivascular spaces (PVSs), which are also known as Virchow-Robin spaces, are cerebrospinal fluid (CSF) filled spaces ensheathing small blood vessels as they penetrate the brain parenchyma. The clinical significance of PVSs comes primarily from their tendency to dilate in abnormal cases. Particularly, substantial research based on dilated PVS has been used for the diagnosis of Alzheimer’s disease, stroke, autism, etc. In general, the PVS structures could be identified using brain magnetic resonance imaging (MRI). Particularly, quantitative analysis of PVSs based on MRI data is important for revealing correlations between cerebrovascular lesions and neurodegenerative diseases. However, since manual delination of PVSs is tedious and time-consuming, automatic and accurate segmentation of PVS is highly desirable in PVS-based studies.
In this study, we propose a structured-learning-based segmentation framework to extract PVSs from high-resolution 7T MR images. Specifically, we integrate three types of vascular filter responses into a structured random forest for classifying each voxel into two categories, i.e., either PVS or background. Also, we propose a novel entropy-based sampling strategy to extract informative samples in the background for training an explicit classification model. Since the vascular filters can extract various vascular features, even thin and low-contrast structures can be effectively extracted from noisy background. Moreover, continuous and smooth segmentation results can be obtained by utilizing patch-based structured labels. The performance of our proposed method has been evaluated on 19 subjects with 7T MR images. The experimental results demonstrate that the joint use of entropy-based sampling strategy, vascular features, and structured learning improves the segmentation accuracy, with the Dice similarity coefficient reaching 66%.