 Austin J. Brockmeier, Jose C. Principe, University of Liverpool, UK, University of Florida, USA
Austin J. Brockmeier, Jose C. Principe, University of Liverpool, UK, University of Florida, USA
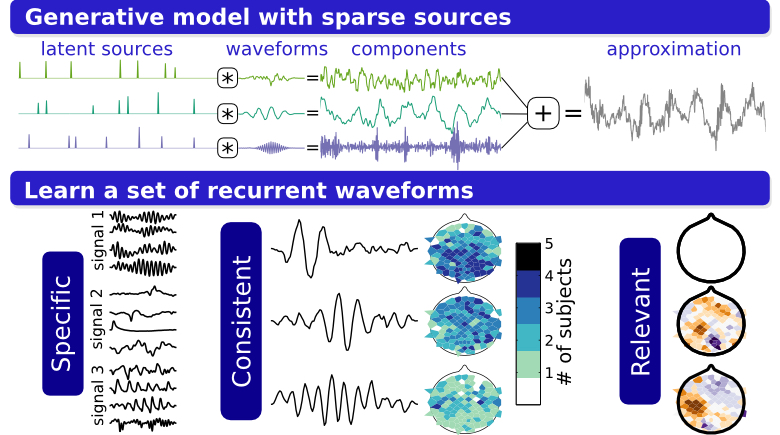
When experts analyze EEGs they look for landmarks in the traces corresponding to established waveform patterns, such as phasic events of particular frequency or morphology. Lengthy records motivate automated analysis techniques, but these are often constrained by design choices that may not match the structure of the EEG signals. This paper explores a modeling approach that automatically learns the waveforms corresponding to transient, reoccurring events within EEG traces. The methodology is based on a sparsely excited model of a single EEG trace, and the model parameters are estimated using shift-invariant dictionary learning algorithms developed in the signal processing community. The contributions of the study include a multistage estimation approach, a protocol for cluster analysis of the waveforms learned across electrodes and subjects, and an application to motor imagery classification using the spatial amplitude patterns associated with the learned waveforms. On the motor imagery dataset, linear discriminant analysis can distinguish the type of motor imagery based on the spatial patterns of a subset of the learned waveforms.
MATLAB code for the methodology is available from the Computational NeuroEngineering Lab (CNEL) at the University of Florida http://cnel.ufl.edu/~ajbrockmeier/eeg/
Keywords: Biomedical signal processing, clustering, dictionary learning, EEG, sparse coding.

