Researchers are cataloging and analyzing every cell type in the brain through the National Institutes of Health’s The BRAIN Initiative®
For over a century, scientists have known that the brain contains multiple cell types, dating back to Santiago Ramón y Cajal’s earliest observations of brain tissue under a microscope. But until recently, we lacked the tools to study those cells with enough resolution to truly understand their roles.
Major advances in both technology and methodology have finally changed that. Researchers are now able to study the molecular fingerprints of brain cells, including how they differ from one another in gene expression—and how they differ from the brain cells of other species. Insights gleaned from these investigations can enhance our overall understanding of how the brain functions; they can also offer critical clues about targeted treatments for mental health disorders and other disorders not seen in other species.
“A big question is what makes us uniquely human,” said Andrea Beckel-Mitchener, Ph.D., deputy director of the National Institutes of Health (NIH) Brain Research through Advancing Innovative Neurotechnologies (BRAIN) Initiative. “The more we learn about human brains and circuitry, the better we’re going to understand what components are unique to us as a species and what potential treatments will be useful in the future.”
A comprehensive cell-type atlas
In 2014, with the ultimate goal of identifying and characterizing all cell types in the human brain, the U.S. NIH funded a series of pilot projects centered on developing methods for studying brain cells across various organisms. That initial phase was followed by the launch of the BRAIN Initiative Cell Census Network (BICCN) in 2017, which focused on characterizing cells in the mouse brain (how they develop, where they are located, and how they coordinate with other cells) and included research on humans and nonhuman primates. In 2023, NIH launched the final stage of the initiative, the BRAIN Initiative Cell Atlas Network (BICAN), which aims to support research across the field of neuroscience by creating a comprehensive reference atlas of cell types in the human brain throughout the lifespan.
One of the key insights so far is that few differences exist in brain cells between humans and nonhuman primates. “It’s actually pretty subtle, but it’s enough to make our ability to think, behave, and predict—those advanced cognitive skills that are apparent in humans are probably due to a relatively small number of gene expression changes,” Beckel-Mitchener said. Differences in gene expression can have a drastic impact on cells, including how they develop, what proteins they produce, and how they respond to other cells and environmental factors.
A BICCN study led by researchers at the Allen Institute for Brain Science used single-nucleus RNA sequencing to profile the gene expression of individual cells, comparing humans to chimpanzees, gorillas, rhesus macaques, and marmosets. The team sequenced more than 570,000 cells from the middle temporal gyrus, a region associated with language abilities, finding that cell types were largely unchanged between species. Differences were found primarily in pathways responsible for communication between neurons and glial cells [1].
“The protein coding part of genes evolves very slowly, and cell types themselves are probably not changing much from one species to the next,” said Jesse Gillis, Ph.D., an associate professor at the University of Toronto’s Donnelly Centre for Cellular and Biomolecular Research and a coauthor of the study. “But the organization of cells, and their gene regulation, is changing.”
The ability to study gene expression at the single-cell level has been a game changer, because cellular functions can shift based solely on their gene expression. Based on insights from single-cell sequencings, cells that may otherwise look the same can therefore be classified as different types. “We can get a snapshot of cells across the entire brain—not just based on their shape or the neurotransmitters they respond to—but their entire genetic makeup,” said Arnold Kriegstein, M.D., Ph.D., a professor of neurology at the University of California, San Francisco’s Weill Institute for Neurosciences.
Gillis and his colleagues are even further amplifying the impact of the new technologies with a clever approach: merging single-cell data with bulk data from the population level. For some species, such as chimpanzees, researchers have access to a limited number of single-cell samples. “We can add another layer of evidence if we can take regulatory changes that we infer from a single cell, then validate them using bulk population data,” Gillis said.
Using that method, he and his colleagues again compared cells from the middle temporal gyrus of humans to those of chimpanzees, gorillas, rhesus macaques, and marmosets (Figure 1). They found recently evolved gene expression differences in 24% of genes studied, many of which were associated with human-specific brain diseases. For example, the human BBS5 gene had significant differences in its regulation compared to the other species studied. That gene is closely linked to Bardet–Biedl syndrome, a rare genetic condition that causes intellectual impairment and problems with multiple systems in the body [2]. Understanding the role of various cell types in that condition could be a key step toward developing targeted treatments, Gillis said.
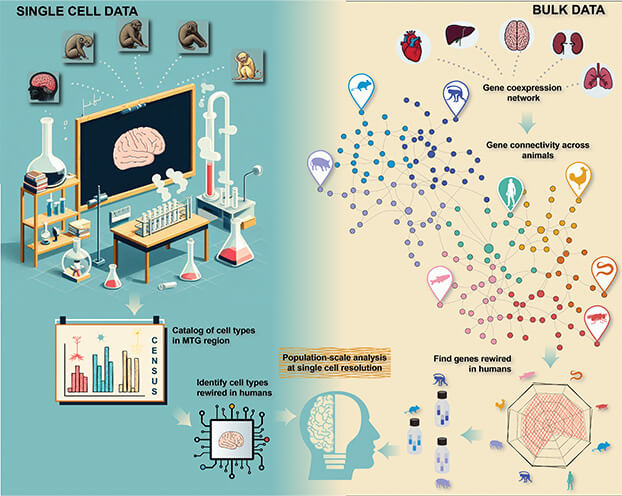
Figure 1. Gillis and his colleagues are working to understand cell-type differences at population scale. Single cell data provides a catalog of cell-types in a small number of individuals from targeted primates (left). Bulk data provides co-expression (gene-gene correlation) using a large number of individuals (right). By combining the two, it is possible to validate cell-type differences in regulation at population scale. (Image courtesy of Jesse Gillis and Hamsini Suresh.)
How brain cells change over time
When it comes to understanding and treating diseases that uniquely affect humans, researchers are not only analyzing cells from the adult brain, but also from brains throughout various stages of development. “We think studying development is particularly important because we know that diseases such as autism and schizophrenia have their onset at very early stages of brain development,” Kriegstein said. As the brain develops, some cell types appear and disappear and regions can change significantly in terms of cellular composition. Those dynamic changes are part of what allows humans to develop complex speech and motor abilities, but they can also make us vulnerable to disease. For that reason, Kriegstein and his colleagues are working to develop a reference database of how brain cell types change over time.
With single-nucleus RNA sequencing, he and his colleagues studied more than 700,000 cells from the human cortex, beginning in very early fetal development and extending into adulthood (Figure 2). They found that in the subplate region, a structure that helps guide the development and wiring of the brain’s cortex, genes associated with autism risk were more highly expressed in female cells than in male cells [3].
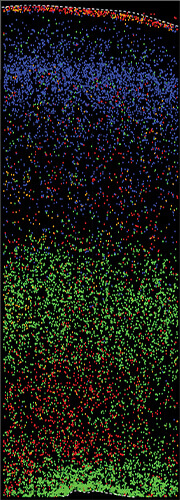
Figure 2. In this image of the developing human cortex, four cell types (each a different color) were identified by gene expression. They occupy distinct cortical layers. Red and orange represent two interneuron subtypes; green represents astrocytes; and blue represents excitatory neurons. (Image courtesy of Tanzila Mukhtar, Kriegstein Lab.)
“The fact that there’s a higher expression of risk genes in females than males may seem contradictory at first, given that autism is much more common in males,” he said. “However, this is consistent with the hypothesis that females are protected in some way against the loss of function in those genes. We think that the higher expression level of these particular cells offers females a protection that males don’t have.” During early stages of development, neurons in the subplate region project to areas of the cortex associated with learning, memory and higher cognitive functions, then shift into deeper parts of the brain over time. The genetic differences Kriegstein and his colleagues observed between male and female autism risk cells suggest that the cellular changes occurring in this region may be a key part of the vulnerability to that disease.
The researchers also studied how differences in gene expression of cells relates to other diseases, including schizophrenia and Alzheimer’s. They observed that each disease’s risk genes were linked to different cell types in different regions of the brain. For example, regulatory changes in microglia, immune cells that help respond to damage and pathogens in the brain, were associated with Alzheimer’s disease vulnerability [3]. Past research has indicated that other neurodegenerative disorders, such as Parkinson’s, are also related to problems with specific types of cells [4]. “We think that having that kind of a map of cell types and which connections they make will eventually help us develop very targeted treatments,” said Beckel-Mitchener.
Fundamental and applicable findings
Building on the breakthroughs of BICCN, the NIH has now launched BICAN, which also leverages multiple innovative single-cell spatial mapping approaches to take cell type analysis one step further. Researchers can now collect single-cell recordings that include data on spatial changes, allowing them to observe how brain cells migrate and connect over time. “This technology is so new that what’s considered the best keeps changing from year to year,” Kriegstein said. “But if you want to take advantage of cutting-edge technology, you’ve got to jump in at some point.”
Some new projects will turn exclusively to the human brain. Kriegstein and his colleagues are conducting single-cell analysis of autism in the brain at various ages. Given what they have learned about the cell types that are most vulnerable during development, can they also observe changes in those types of cells as a physical manifestation of the disease itself? Gillis and his team are amassing samples from hundreds of people to begin studying subtle variability in cell-type gene expression among humans.
Other work will use the latest technologies and methodologies to dive deeper into cross-species comparisons. Comparing the brains of humans, macaques, and mice, Kriegstein has shown that human synapses remain immature (and therefore more plastic) for an extended period, which helps explain our increased capacity for learning and memory [5]—and aims to further explore such developmental distinctions. Gillis is continuing to use single-cell recordings to study cell-type differences in a wide variety of organisms, including everything from plants to armadillos. “Finally, at cellular resolution, we can really align the data and compare species,” he said. “That allows us to start building out cellular atlases and makes integrative assessments more of an apples-to-apples comparison.”
Much of the research from BICCN and BICAN is fundamental in nature, offering knowledge about some of the smallest units of brain function. But findings on disease-linked cell types is already revealing targets for specialized therapies that could make the research applicable on a relatively short timeline. “This program was structured so that it could support institutes across NIH with more specialized missions,” Beckel-Mitchener said. “We’re already starting to see the excitement from many different research communities about these revolutionary findings on cell types.”
References
- N. L. Jorstad et al., “Comparative transcriptomics reveals human-specific cortical features,” Science, vol. 382, no. 6667, Oct. 2023, Art. no. eade9516, doi: 10.1126/science.ade9516.
- H. Suresh et al., “Comparative single-cell transcriptomic analysis of primate brains highlights human-specific regulatory evolution,” Nature Ecol. Evol., vol. 7, pp. 1930–1943, Sep. 2023, doi: 10.1038/s41559-023-02186-7.
- D. Velmeshev et al., “Single-cell analysis of prenatal and postnatal human cortical development,” Science, vol. 382, no. 6667, Oct. 2023, Art. no. eadf0834, doi: 10.1126/science.adf0834.
- J. Bryois et al., “Genetic identification of cell types underlying brain complex traits yields insights into the etiology of Parkinson’s disease,” Nature Genet., vol. 52, pp. 482–493, Apr. 2020, doi: 10.1038/s41588-020-0610-9.
- L. Wang et al., “A cross-species proteomic map reveals neoteny of human synapse development,” Nature, vol. 622, pp. 112–119, Sep. 2023, doi: 10.1038/s41586-023-06542-2.