The most advanced diagnostic technologies may be amazing, but they often do not make it to resource-limited settings, including those places where particularly dangerous pathogens are more prevalent, crowded conditions make outbreaks more likely, or medical facilities are less available.
“The main problem is access to health care, and the farther you get from higher resource settings, the harder this is to deliver,” said Keith Pardee (Figure 1), Ph.D., Canada Research Chair in Synthetic Biology in Human Health and an associate professor in the Leslie Dan Faculty of Pharmacy at the University of Toronto.

Figure 1. Keith Pardee (fourth from left), the Canada research chair in synthetic biology in human health, and an associate professor in the Leslie Dan Faculty of Pharmacy at the University of Toronto, with his research group and collaborators from Brazil, Colombia, and Ecuador while hiking to the Pichincha Volcano above Quito, Ecuador. Pardee’s group is developing highly accurate but inexpensive diagnostic technology, particularly as a way to reach resource-limited settings. (Photo courtesy of Keith Pardee.)
Pardee’s group, along with that of Alexander Green (Figure 2), Ph.D., associate professor of BME at Boston University, have collaborated on an innovative technology that identifies pathogens rapidly and affordably, and with an accuracy that rivals the highly accurate but far more expensive quantitative reverse transcription polymerase chain reaction (RT-qPCR) test for detecting viral infection. Their technology works by identifying a target pathogen, which triggers production of a protein that yields one of several easily readable signals, including one that can be read by a run-of-the-mill blood-glucose meter.

Figure 2. Alexander Green, an associate professor of BME at Boston University, whose research group often collaborates with that of Pardee on innovative, cell-free, toehold-switch systems to identify pathogens rapidly and affordably. Beyond that work, Green’s group is also beginning a project, funded by the Defense Advanced Research Projects Agency (DARPA), to develop a similar 30-minute test to identify key biomarkers that determine a person’s readiness to perform either a physical or cognitive task. (Photo courtesy of Qing Hua Wang.)
“When you think about things like the Zika virus, Ebola, and SARS-CoV-2, there’s a clear need in the world for an ability to detect viruses very sensitively and without a lot of instrumentation,” Green said. “My lab had already been working on engineering RNA to detect different species, so this seemed like a good opportunity to address something that could help a lot of people.”
Green and Pardee are two of four co-founders of a company—En Carta Diagnostics, headquartered in Paris, France [1]—that is now advancing the next-generation technology to market, with an emphasis on speed, cost, and ease of use for nonskilled users.
Identifying pathogens
A core piece of the detecting approach is the toehold switch. Toehold switches are pieces of RNA that operate within in vitro biochemical reactions and incorporate binding regions of RNA, which are complementary to a pathogen genome. When a binding region links to, or gets a “toehold,” on the target pathogen, it switches on a synthetic gene circuit to allow the ribosome to bind to the toehold switch and begin the process of making a chosen signaling molecule [2], Green explained.
To get this to work, he and his collaborators manipulated how an RNA molecule interacts with the ribosome to initiate translation. “Based on knowledge of how this process works, we were able to engineer the RNA molecule to work as a switch that regulates when an mRNA is being translated, ensuring that it only turns on translation when it binds to a target sequence,” he described. From there, the mRNA yields the chosen protein.
Green and his research group have now designed improved toehold-switch systems that not only detect the virus, but also generate a different signal for one mutation of the virus versus another [3]. “You can get more specific detection that way,” Green said. His group has also begun developing biochemical systems to identify the presence of nonnucleic acids, such as chemical water contaminants.
Low-cost reporting of pathogens
Numerous methods may be used to report identification of a pathogen, and the researchers have developed several economical options. One involves embedding the cell-free, toehold-switch system on a piece of paper, and freeze-drying the paper so it can be stored for at least a year with no need for refrigeration [4]. This work, done by Pardee’s group, represents a significant improvement over typical cell-free, protein-synthesis systems that must be stored at minus 80 °C to retain their activity.
The first generation of this technology used paper disks that were 2 mm in diameter, and read the signal output with a compact, battery-operated, simple-to-use plate reader that his students built for $500, and reported easy-to-read results (Figure 3). This compares to standard PCR machines that are far larger and cost 60 times more, Pardee described. Much of the cost in PCR lies in the thermal cycling reaction that requires 30–40 rounds of precise heating and cooling to amplify the target RNA to sufficient quantities for molecular and genetic analyses. The paper-based system likewise requires amplification for the toehold switch to do its analysis, but can do so with isothermal (one-temperature) methods instead.
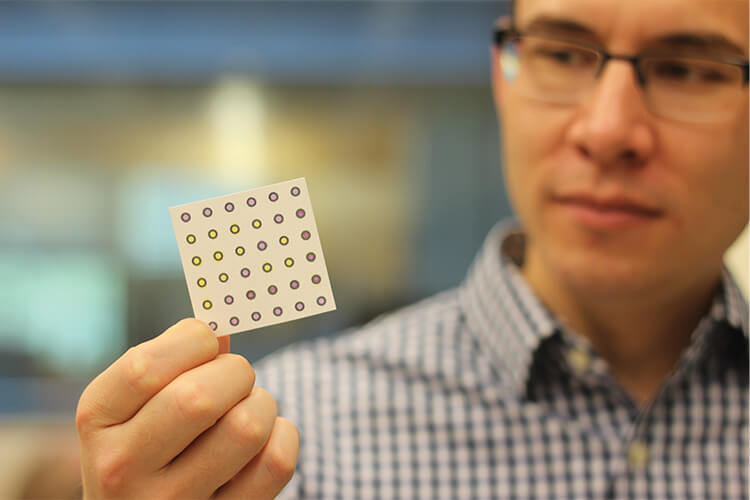
Figure 3. Green holds an example of a project developed through collaboration with Pardee’s group. It is an assay that makes use of the cell-free system, which is embedded in a paper substrate; combines it with rapid isothermal nucleic acid amplification reactions; and ultimately displays color changes (shown) to indicate whether patient samples are infected with target viruses. (Photo courtesy of Qing Hua Wang.)
After demonstrating their system worked, the members of Pardee’s group then began thinking about other protein outputs and how to read them. One of those group members was doctoral student Evan Amalfitano (Figure 4), who used a toehold switch to activate glucose production, and then employed a standard blood glucose reader to display the results (Figure 5). “Glucose meters are great examples of very low cost and accessible electrochemical sensors: the glucose meter strips are a few cents each, it’s a very well-established technology, and the meters themselves are $15–$30 (USD).”

Figure 4. Evan Amalfitano, postdoctoral researcher and previous doctoral student in the Pardee lab, devised a test that uses a toehold switch to activate glucose production. (Photo courtesy of Steven Southon.)

Figure 5. Amalfitano’s test employed a standard blood glucose reader to display his test results. An incubator (the black box) was developed by other students in Pardee’s lab to amplify the target’s genetic signature so the toehold switch can detect it, and then carry out the toehold-switch reaction itself. Amalfitano is now collaborating with the National Research Council Canada to advance that technology. (Photo courtesy of Steven Southon.)
For months afterward, Amalfitano struggled to find an enzyme that would output glucose, finally settling on trehalase. “I had never heard of trehalase before, but it was available, so I tried it out and it worked amazingly well within our system,” he said. Tests on Zika, COVID-19, typhoid (a bacterial target), and other infections showed the glucose system accurately identified them all [6]. He remarked, “That’s one of the really nice things about the toehold switch: it can be adapted to detect almost anything.”
Their tests also showed that the system could multiplex. “You can add at least two different targets to the same reaction, and see different glucose levels that indicate which toehold switch is activated,” Amalfitano said. They showed the multiplexing capability with typhoid fever and an antibiotic resistance that is often associated with typhoid fever. “We could see if it was positive for typhoid, and if so, whether there was antibiotic resistance,” he said, noting that such information is important when selecting treatment options.
Going forward
Amalfitano, who is now a postdoc in Pardee’s group, is currently further developing the glucose sensor technology at the National Research Council Canada (NRC) facility in Boucherville, Quebec. “What we’re working on now with the NRC is trying to automate that whole process, and we have developed a chip that contains all the reagents for the reaction, so we can put the patient sample on the chip, and put the chip into a microfluidic device that mixes the reagents and incubates them as needed to carry out the isothermal amplification and toehold-switch reaction.”
Next up, he would like to integrate the glucose-reading into the automatic system. “If we could do that, you’d just add your sample to the chip and it would automatically do the measurement and give you the results,” Amalfitano asserted.
For Pardee, speed and automation will make the tests more useful. In 2022, he and his group, along with Green, published field validation of their paper-based tests for Zika and chikungunya viruses in more than 300 patient serum samples: 268 samples for Zika, and 65 for chikungunya [7]. “We showed just over 98% accuracy compared to RT-qPCR, and while we were happy with that, the assay took two to two-and-a-half-hours, so there was need for improvement. This is where the team and En Carta are now focused,” he said.
Similar to the Amalfitano/NRC project, En Carta is planning to develop a chip to measure a sample and get the output automatically, Green said. “The chip would heat isothermically for 30 minutes or so before cooling down, and over that time, the reaction occurs, the sensors bind, and you get the readout.” Meanwhile, Pardee’s group at the University of Toronto is gearing up for a large patient trial of the new system, which will be conducted in four or five countries and with three or four pathogens.
As that proceeds, Green and his Boston University lab are continuing to collaborate with Amalfitano and Pardee by providing new RNA and DNA sensors for various toehold-switch diagnostics, and are working on some cell-free, fluorescent-reporting diagnostics of their own.
For Pardee and his group, they are beginning to expand their work from diagnostics to pathogen therapeutics. “Distributing the diagnostics is one big challenge, and the next is distributing the therapeutics,” Pardee said. “At the moment, mRNA vaccines need refrigeration to minus 80 °C, and antigen-based vaccines also need refrigeration. We need to find better alternatives. This is an active area for us and for others in the research community, because as medicine moves more and more toward molecular solutions, cold storage will be an increasingly important problem.”
Solving the access problem
For now, the primary objective of Pardee, Amalfitano, and Green is the same: making clinical-grade diagnostics available to everyone. That means learning first-hand what the access issues are and what will work in resource-limited settings (Figure 6) ranging from low- and middle-income nations to remote areas within developed countries and poorer neighborhoods in large urban centers, Pardee said. “It’s why we have meetings every two weeks with international research partners who serve these settings, why we’ve been hosting students here in our lab and sending our students there,” he remarked. “In-person exchanges are absolutely priceless in terms of technology transfer and just making sure that you’re building something robust and resilient for the places that need better options.”

Figure 6. Pardee’s team collaborates with researchers and others in resource-limited settings to ensure that the diagnostic technology they develop is well suited to the region. This includes face-to-face meetings, such as this visit to Ecuador. (Photo courtesy of Keith Pardee.)
Effective, fast, easy-to-use, and inexpensive diagnostic technologies are imperative, Green agreed. “I do think we’re heading toward a time when people can get gold-standard, PCR-level assays literally anywhere, and when reaction components will be in a chip so anyone can apply a sample—even in an at-home test—and get easily readable test results in under 30 minutes.” He added, “We need ubiquitous detection technology so we not only know where pathogens are coming from and when they emerge, but also so we can help protect people in resource-limited areas, where their biggest problem is they don’t have enough money. It shouldn’t be like that. It’s definitely an unmet need.”
References
- En Carta Diagnostics. Accessed: Feb. 13, 2024. [Online]. Available: https://www.encarta.bio/
- A. A. Green et al., “Toehold switches: De-novo-designed regulators of gene expression,” Cell, vol. 159, no. 4, pp. 925–939, Nov. 2014.
- D. Ma et al., “Multi-arm RNA junctions encoding molecular logic unconstrained by input sequence for versatile cell-free diagnostics,” Nature Biomed. Eng., vol. 6, no. 3, pp. 298–309, Mar. 2022.
- K. Pardee et al., “Paper-based synthetic gene networks,” Cell, vol. 159, no. 4, pp. 940–954, Nov. 2014.
- K. Pardee et al., “Rapid, low-cost detection of Zika virus using programmable biomolecular components,” Cell, vol. 165, no. 5, pp. 1255–1266, May 2016.
- E. Amalfitano et al., “A glucose meter interface for point-of-care gene circuit-based diagnostics,” Nature Commun., vol. 12, no. 1, p. 724, Feb. 2021.
- M. Karlikow et al., “Field validation of the performance of paper-based tests for the detection of the Zika and chikungunya viruses in serum samples,” Nature Biomed. Eng., vol. 6, no. 3, pp. 246–256, Mar. 2022.



