Sepsis is an urgent medical problem around the world with at least 47 million affected per year. Of those, about a quarter—at least 11 million—die, and many sepsis survivors face health repercussions for the rest of their lives, according to the Global Sepsis Alliance [1]. Along the same lines, the World Health Organization named sepsis as the cause of one in five deaths globally, and the U.S. Centers for Disease Control and Prevention (CDC) notes similiarly dire statistics [2]. These are some of many dire statistics that have prompted health agencies and biomedical innovators to address this massive health emergency.
A major challenge to fighting sepsis is the speed of its attack. It can escalate to severe organ dysfunction and death in a matter of hours, which means clinicians must correctly diagnose it and start proper treatment very quickly. To address sepsis response in medical institutions, the U.S. Centers for Disease Control and Prevention has just launched an effort to work even more pointedly with hospitals to hone their sepsis programs. On another front, researchers have been busy developing rapid and definitive diagnostic tests, as well as high-speed technologies to identify which of the wide range of pathogens underlies each patient case so the proper treatment can begin.
Beefing up efficiency
“The CDC has been active in trying to improve the science around sepsis, and trying to improve early recognition and awareness of sepsis for many years now. The difference this year is that we published the Hospital Sepsis Core Elements [3],” said Raymund Dantes, M.D., MPH, CDC medical advisor and lead sepsis expert (Figure 1). Dantes is also an associate professor of medicine at Emory University School of Medicine in Atlanta, Georgia.

Figure 1. Raymund Dantes, M.D., MPH, CDC medical advisor and lead sepsis expert, as well as associate professor of medicine at Emory University School of Medicine in Atlanta, Georgia. (Photo courtesy of Emory University.)
While plenty of evidence-based and regularly updated clinical guidelines already exist for the detection and treatment of sepsis, something was missing, according to Dantes. “Our Morbidity and Mortality Weekly Report (MMWR) [4] found that three-quarters of hospitals have a sepsis program or sepsis committee already in place, but that means there are more than a thousand hospitals that do not,” Dantes said (Figure 2). “The Core Elements are about trying to help those hospitals get their sepsis programs off the ground, and for those hospitals that already have sepsis programs, giving them ideas about how they can improve their sepsis care as well.”
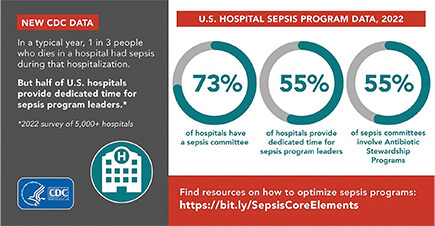
Figure 2. Data from the CDC shows the great health impact of sepsis. (Image courtesy of the CDC.)
In addition to the establishment of carefully thought-out sepsis programs, the Core Elements urges hospitals to officially assign a program leader who has dedicated time to record and track sepsis response and care. That information can then be used to identify and draw attention to opportunities for betterment, and potentially make a case to hospital administrators for remedy, Dantes said, especially as timely sepsis care becomes part of U.S. Centers for Medicare and Medicaid Services hospital incentive payment programs in 2024.
Beyond the Core Elements effort, Dantes believes new technologies are poised to save lives too. For instance, he said, groups are developing machine-learning and artificial-intelligence tools to mine electronic health records and provide predictions of a patient’s risk of developing sepsis, and others are designing a variety of tests to help clinicians detect sepsis and begin treatment earlier. He remarked, “It’s all really exciting stuff that’s going on.”
New sepsis test
In terms of sepsis tests, the FDA just cleared a diagnostic tool developed by Cytovale of San Francisco, CA, and designed to pull information from a standard-type blood draw, run it through a computer algorithm, and determine a patient’s risk of developing sepsis. And it does it all in 10 minutes, according to biomedical engineer Carissa Colegrove (Figure 3), associate director of product development for Cytovale.
Figure 3. Biomedical engineer Carissa Colegrove, associate director of product development at the medical diagnostics company Cytovale. (Photo courtesy of Cytovale.)
“Sepsis is basically an infection that has gone out of control, and early research showed that there was good mapping between the deformability of white blood cells and indications for sepsis,” Colegrove said (video at https://vimeo.com/546722700/a4afaf9702). The reason for this deformation is based on the concept that white blood cells expel proteins while fighting an infection, making the cell walls less rigid [6]. Cytovale capitalized on the connection. “We had to go from a theory that says (septic) white blood cells squish more and healthy white blood cells squish less, run many different studies over time, and train an algorithm to be able to tell the difference with meaningful clinical performance,” she described.
The algorithm was just one of three major challenges they needed to solve. A second was a making a microfluidics chip capable of lining up white blood cells one by one, so each could be compressed in exactly the same way every time, she said. “That, in and of itself, was pretty new technology, and then going from prototypes to mass manufacturing was another whole part of the journey.”
The third major challenge was building an instrument that could execute multiple functions, she explained. That included sample preparation, or separating the white blood cells from the whole-blood draw; alignment of the microfluidics chip to within 0.3 microns so it sits precisely over a camera that is then able to capture a video of the cell as it is being compressed; and delivery of the video image data to the algorithm for processing, Colegrove explained.
“When I started working at Cytovale, they had their microfluidics more or less dialed in and they had the first versions of the algorithm,” Colegrove recalled, “but while they had an instrument that worked well enough to collect the clinical data, it was more like a hobbyist’s—pieced together with off-the-shelf items. My job over about five years was to figure out how to take this big instrument that takes engineers to run, and turn it into a commercial product that fits on a lab bench and can return results by itself.” She and her team succeeded. The instrument now combines a 1 m × 1 m × 1 m cell-imaging module with an adjacent 0.5 m × 0.15 m sample-prep module, “so the whole thing takes up about half a lab bench” (Figure 4).
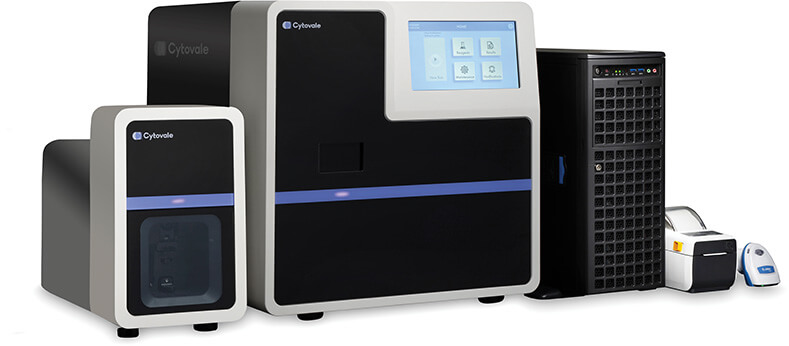
Figure 4. Cytovale’s IntelliSep system combines a multifunction instrument, a specially designed microfluidics chip, and an algorithm that together process and analyze the deformability of white blood cells to quickly determine a patient’s risk of developing sepsis. Now FDA-cleared, the first system is already in use in a large U.S. hospital’s emergency room, and the company plans to expand to other hospitals. (Photo courtesy of Cytovale.)
A multicenter study [7] showed that the system, called IntelliSep, had a 97% negative predictive value and a 50% positive predictive value in determining likelihood for sepsis. In other words, she said, among those patients that the test determined had a low risk of sepsis, 97% did not develop sepsis; and among those determined as high risk, 50% did go on to develop sepsis.
Following the FDA clearance in December 2022, Cytovale has begun rolling out IntelliSep to hospitals. The 1,000-plus-bed Our Lady of the Lake Regional Medical Center in Baton Rouge, Louisiana, became the first to adopt the technology for use in its emergency department. “The idea is to start at one hospital, expand across that hospital system (which has locations in Mississippi and Louisiana), and then go from there,” Colegrove said. She anticipates a warm welcome by clinicians, because the system not only provides sepsis risk quickly, but it requires no extra effort on the part of clinicians. “They don’t even have to collect the blood sample a different way. All they have to do is say, ‘Run that test on the sample, too,’” she said. The results are also user-friendly, with a score of 0.1 to 10 and results separated into one of three bands, where band 1 is low likelihood of developing sepsis in the next three days, band 3 is high likelihood of developing sepsis in the next three days.
Cytovale’s long-term goals include marketing IntelliSep to global medical facilities, and looking into other areas where this technology could be applicable, she said. “For the next three years, however, scale-up of our current product in the U.S.—making a bunch of them and supporting them—is our main focus.”
Host-microbe metagenomic sequencing
An example of a group using machine-learning technologies is that of Chaz Langelier (Figure 5), M.D., Ph.D., associate professor of medicine in the University of California San Francisco Division of Infectious Diseases, and an investigator at the Chan Zuckerberg Biohub in San Francisco. “This particular type of technology is called host-microbe metagenomic sequencing. Essentially, it involves profiling not only the DNA and RNA from microbes in blood samples, but also the patterns of gene expression that exists in those samples,” he said. “It captures both the microbe causing the problem and the host immune response, which is massively dysregulated in sepsis.”

Figure 5. Chaz Langelier, M.D., Ph.D., associate professor of medicine in the University of California San Francisco Division of Infectious Diseases, and an investigator at the Chan Zuckerberg Biohub in San Francisco. (Photo courtesy of Chaz Langelier.)
His group initially worked with computational biologists Katrina Kalantar, Ph.D., of the Chan Zuckerberg Initiative, and Lucile Neyton, Ph.D., of UCSF, to develop a similar approach to improve the diagnosis of pneumonia. “That was really motivated by the fact that pneumonia is very challenging to diagnose in many critically ill patients, because a lot of other noninfectious processes can look like pneumonia,” he said. “We used machine-learning approaches on metagenomic data from the host immune side and the microbial side, and built models that could accurately distinguish whether patients had pneumonia or noninfectious acute respiratory failure in the intensive care unit.” The result was an assay that could simultaneously profile the host immune response and identify a signature of infection, while also detecting the microbial changes that are characteristic of pneumonia [8].
To translate the pneumonia technology to sepsis, Langelier and his group determined how to detect microbes not in respiratory fluid, but in plasma samples. A primary issue was that plasma contains a very low concentration of microbial nucleic acid, so they had to also ensure that the sequencing was picking up nucleic acid from the microbe and not from contamination stemming from the surrounding laboratory environment, he explained. “We needed to undergo a number of iterations, including sequencing in a clean room, before we were confident that what we were detecting were truly microbial sequences from the blood of patients.”
They also had to develop an algorithm for sepsis. For that, they accessed data gathered from blood and plasma samples of more than 300 critically ill patients that had been admitted to the UCSF Medical Center and Zuckerberg San Francisco General Hospital between 2010 and 2018, and trained the algorithm using a retrospective assignment of sepsis for each patient based on traditional clinical determination.
Concurrently, they employed metagenomic next-generation sequencing to compare the pathogen’s genetic data against reference genomes. This combination allowed them to identify 99% of microbiologically confirmed sepsis cases, and predict sepsis in 74% of suspected and 89% of indeterminate sepsis cases, and also determine the pathogen, whether it be bacterium, virus, or fungus [7]. Such information is critical, because the treatment for sepsis—including suspected but not confirmed sepsis—is delivery of antibiotics, Langelier said, “and in 30%–65% of cases, there’s never a microbe that’s identified despite the best efforts of the medical team in the hospital. That means the antibiotics may not be optimally matched to the microbe causing the problem.” Delivery of antibiotics to nonseptic patients is also an issue, because unneeded antibiotics may make the patient more vulnerable to other opportunistic hospital infections, such as Clostridioides (Clostridium) difficile, and can potentially delay patients from getting the timely treatments they actually do need.
The next step for the Langelier group’s work is independent validation of their results in a separate multicenter cohort of patients, and also in pediatric patients, and then the start of clinical validations. CZ Biohub and UCSF have already filed a provisional patent on the technology, and have initiated discussions with companies interested in developing it further.
While that is progressing, Langelier is working on related research. “It turns out that when you’re sequencing the genomes of microbes, you can also detect the genes that cause antibiotic resistance. In fact, we had a study last year [10] that demonstrated we could predict with reasonable accuracy—and without a need for culture—the antimicrobial-resistant profile of patients with drug-resistant bacterial pneumonia,” he said. “We’re working now to improve those models, as well as to extend that work to detect antimicrobial-resistance genes belonging to sepsis pathogens in the blood.”
He and his group are also exploring the use of the technology to predict outcomes, such as survival rate at 30 days, he noted. “If we can identify the patients who are at the high at risk of dying from sepsis, then that could open the possibility for more aggressive interventions to prevent adverse outcomes and to ensure that they get the maximal care possible.” In addition, he and his group are also using the approach of simultaneously profiling host and microbe to gain a better understanding of the pathophysiology of sepsis, pneumonia, and other infectious and inflammatory diseases. “For instance, we’re looking into the immune response to sepsis and pneumonia, how it changes with age, and whether we can identify specific therapies that could be more effective in older adults.”
Langelier added, “We urgently need better diagnostic tests for sepsis and for pneumonia, and I think this technology could really make an impact.”
References
- Global Sepsis Alliance, Webpage, Centers for Disease Control Prevention. Sepsis Facts. Accessed: Oct. 9. 2023. [Online]. Available: https://www.worldsepsisday.org/sepsisfacts
- World Health Organization. Global Report on the Epidemiology and Burden of Sepsis: Current Evidence, Identifying Gaps and Future Directions. Accessed: Oct. 14, 2023. [Online]. Available: https://iris.who.int/bitstream/handle/10665/334216/9789240010789-eng.pdf
- Hospital Sepsis Program Core Elements. Accessed: Oct. 9, 2023. [Online]. Available: https://www.cdc.gov/sepsis/core-elements.html
- R. B. Dantes et al., “Sepsis program activities in acute care hospitals—National healthcare safety network, United States, 2022,” Morbidity Mortality Weekly Rep., vol. 72, no. 34, pp. 907–911, Aug. 25, 2023, doi: 10.15585/mmwr.mm7234a2.
- Centers for Medicare and Medicaid Services, Fact Sheet. (Aug. 1, 2023). FY 2024 Hospital Inpatient Prospective Payment System (IPPS and Long-Term Care Hospital Prospective Payment System (LTCH PPS) Final Rule—CMS-1785-F and CMS-1788-F Fact Sheet. Accessed Oct. 9, 2023. [Online]. Available: https://www.cms.gov/newsroom/fact-sheets/fy-2024-hospital-inpatient-prospective-payment-system-ipps-and-long-term-care-hospital-prospective-0
- M. G. Sorrells et al., “Biophysical changes of leukocyte activation (and NETosis) in the cellular host response to sepsis,” Diagnostics, vol. 13, no. 8, p. 1435, Apr. 2023, doi: 10.3390/diagnostics13081435.
- H. R. O’Neal et al., “Assessment of a cellular host response test as a sepsis diagnostic for those with suspected infection in the emergency department,” Crit. Care Explor., vol. 3, no. 6, p. e0460, Jun. 2021, doi: 10.1097/cce.0000000000000460.
- C. Langelier et al., “Integrating host response and unbiased microbe detection for lower respiratory tract infection diagnosis in critically ill adults,” Proc. Nat. Acad. Sci. USA, vol. 115, no. 52, pp. 12353–12362, Dec. 2018, doi: 10.1073/pnas.1809700115.
- K. L. Kalantar et al., “Integrated host-microbe plasma metagenomics for sepsis diagnosis in a prospective cohort of critically ill adults,” Nature Microbiol., vol. 7, no. 11, pp. 1805–1816, Oct. 2022, doi: 10.1038/s41564-022-01237-2.
- P. H. Serpa et al., “Metagenomic prediction of antimicrobial resistance in critically ill patients with lower respiratory tract infections,” Genome Med., vol. 14, no. 1, p. 74, Jul. 2022, doi: 10.1186/s13073-022-01072-4.