It was six years ago that fecal transplantation first received prominent media attention and the public began to fully appreciate that the bacteria and other microbes in their bodies could have a real impact on health.
“In that case, the patient was in really bad shape,” recalls Janet Jansson, chief scientist for biology in the Earth and Biological Sciences Directorate at the Pacific Northwest National Laboratory (Figure 1). “She had lost a lot of weight, she was restricted to wearing a diaper, and they had tried every kind of antibiotic combination, but she still couldn’t be cured. This was pretty much a last-ditch effort to cure her.”
Jansson was a key researcher involved in profiling the woman’s microbial community before and after the transplantation. “What we saw was amazing,” Jansson continues. “Her microbial community was really not healthy before the transplantation, but afterward, she immediately assumed the donor’s microbiome. For me, as a microbial ecologist, it was a wonderful and rather unusual example of introducing a microbial community into a new environment, where it was able to establish and have a major impact.”

Since then, fecal transplantation has been used very successfully as a treatment for infections with the bacterium Clostridium difficile, and today many researchers are building their knowledge base to take on the next microbiome-related project to fight a more pervasive illness: inflammatory bowel diseases, or IBDs, which include Crohn’s disease. According to the U.S. Centers for Disease Control and Prevention, IBD affects at least 1 million people in the United States [1].
Omics and Dynamics
Jansson is one of those researchers. Her interest is in investigating the dynamics of the disease using omics technologies [2]. She explains, “We’re combining DNA sequencing, which gives you information about who’s there, the transcripts to get information about what genes are expressed as well as which of those genes are translated into proteins, and the metabolite signatures to get information about the chemical communication within the environment.” Her group applied proteomics and metabolomics techniques for this purpose and collaborated with bioinformaticians (including Robert Knight and Elhanan Borenstein; see the article “Deciphering the Mysterious Microbiome”) to develop algorithms and computational methods to integrate the data.
Already, she and her group have published several interesting findings about the difference in the microbiome of IBD patients compared to that of healthy individuals; they are currently investigating the dynamics of the microbiome composition in patients with IBD. If they can figure out what causes differences in the microbiome, Jansson thinks that knowledge would be useful in targeting future therapies. In the meantime, she is working with gastroenterologists on a more immediate application designed to use simple stool samples from patients to identify fluctuations and potentially predict when a patient might have a symptom flare-up, which can be quite debilitating. This could help both doctors and patients manage the disease.
Beyond IBD, Jansson and her group are also studying the impact of resistant starch (provided by a high-fiber diet) on the gut microbiome. For this project, they have found that individuals on a high-fiber regimen have more of certain beneficial microorganisms in their microbiomes than individuals who are not on such a diet. “Using our multi-omics tools, we can see increases in the types of proteins that those beneficial organisms are producing, as well as metabolites, and get a better idea of how the diet impacts the microbiome. Ultimately, the hope is that you could have a designer diet for different people based on the type of microorganisms they have in their guts.”
Gut-on-a-Chip

A different approach to the microbiome comes in the form of the gut-on-a-chip, originally developed in 2012 [3] by a research group led by Donald E. Ingber (Figure 2, right), a cell biologist and bioengineer and the founding director of the Wyss Institute for Biologically Inspired Engineering at Harvard University. The chip recapitulates the flow and peristalsis-like motions of the human intestine, allowing added human epithelium to spontaneously form intestinal villi with proliferative cell crypts (intestinal glands) and mucus-secreting cells, thereby providing a simulated human gut ripe for experimentation.
Next, Ingber and his team sought to grow both normal microbiome and pathogens on the chips. In December 2015, they announced that they had created a gut-on-a-chip model of human intestinal inflammation and bacterial overgrowth characteristic of IBD [4]. They chose IBD because it has multiple contributing factors, including pathogenic bacteria, immune cells, the microbiome, and the peristalsis-like motion. “It’s very hard to tease out which is important,” Ingber says.
With the gut-on-a-chip, however, the researchers could control each factor individually; and, with the addition of high-resolution imaging, he continues, “we literally have a window on molecular-scale activities inside living human cells in an organ context.” Ingber and his group showed how each factor contributes to the breakdown of the epithelial barrier, loss of villi, production of inflammatory molecules, and other consequences of the disease. They also made the surprising discovery that cessation of peristalsis-like mechanical deformations is a direct cause of bacterial overgrowth, as can be observed in patients with postsurgical ileus as well as IBD.
“In addition, we showed that a normal microbiome—and for that we used a commercial formulation of eight different probiotic bacteria—could actually protect partially against infection by invasive E. coli bacteria,” Ingber explains. More generally, this work demonstrated that it’s possible to conduct extended studies of how the microbiome and normal human cells and tissues interact and what happens when infections and illnesses intervene. “This is an opportunity to go beyond correlations and comparing the microbiome in one patient to another, and to study molecular and cellular mechanisms,” he says, noting that the gut-on-a-chip has been commercialized through a company [5] that is working on applications with pharmaceutical firms and hospitals and has begun exploring its potential for personalized medicine.
Engineering Bacteria

As Ingber’s more-than-50-member research group continues to study the microbiome and develop further organs-on-a-chip, other researchers are leveraging the gut version in their own labs. One of them is Wyss core faculty member Pam Silver (Figure 3, right). Her work involves building on engineered bacteria developed in 2014 to sense exposure to antibiotics [6]. She and her group are now creating a hypersensitive diagnostic to report on whether inflammation is present in the gut as well as where and when the inflammation started.
“We’re using biomarkers of inflammation, specifically a molecule produced by gut bacteria in response to reactive oxygen species made by inflammatory cells. We can suss out whether the engineered bacteria have been exposed to that molecule,” Silver says. She and her group are also developing similar diagnostic molecules for other kinds of health conditions and for dietary purposes (Figure 4). “For example, we have bacteria that can register the level of iron that’s present,” she explains.
Silver’s group has tested its engineered bacteria on animal models and is just beginning to analyze their behavior on the gut-on-a-chip. She thinks this will be an ideal test bed to mimic what goes on in the human gut. Instead of dealing with the confounding assortment of bacteria in a person’s microbiome, it will be possible to build “a totally defined consortia of bacteria on the gut-on-a-chip, where we know every member and its contribution to the consortia, and can manipulate the system in very selective ways.” Besides providing insight into each member’s individual role, she notes, experiments could also determine members’ combined functions.
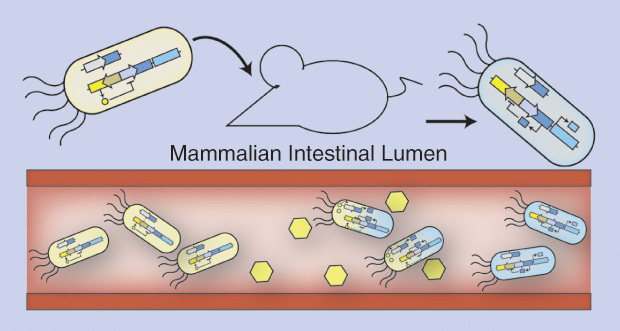
The next step, although admittedly “barely beyond a thought experiment” at this point, is to engineer the bacteria to diagnose an infection and then produce and release a selective or “designer” antibiotic to kill that explicit pathogenic bacterium, Silver says. “Going one step further, there are molecules that could be used to treat inflammatory cells, so we could perhaps engineer bacteria to make those and secrete them. In the same way, it’s believed that there’s a connection between the gut and the brain, and so if we can have the bacteria produce potential neuropeptides or neuromolecules, it could be a way of regulating behavior.”
Like Silver, Jansson is impressed with the number of disease phenotypes potentially tied to the microbiome. Jansson says “Links to autism have been published recently, links to some kinds of cancers, links to rheumatism.” She is even studying the influence of the gut microbiome on the brain through a project funded by the Office of Naval Research.
Still, IBDs are the perfect place to start because they are not only prevalent in the United States but are also becoming more common in other countries that are adopting the Western diet, Jansson says. “These are huge diseases that really impact the people who have them, so it’s very important to understand more about them and their connection to the microbiome.”
References
- U. S. Centers for Disease Control and Prevention. (2015, Mar.) Epidemiology of the IBD. [Online].
- C. Noecker, A. Eng, S. Srinivasan, C. M. Theriot, V. I. B. Young, J. K. Jansson, D. N. Fredricks, and E. A. Borenstein, “Metabolic model- based integration of microbiome taxonomic and metabolomic profiles elucidates mechanistic links between ecological and metabolic variation,” mSyst., vol. 1, no. 1, pp. e00013–e00015, Jan.–Feb. 2015.
- H. J. Kim, D. Huh, G. Hamilton, and D. E. Ingber, “Human gut-on-a-chip inhabited by microbial flora that experiences intestinal peristalsis-like motions and flow,” Lab Chip, vol. 12, no. 12, pp. 2165–2174, June 2012.
- H. J. Kim, H. Li, J. J. Collins, and D. E. Ingber, “Contributions of microbiome and mechanical deformation to intestinal bacterial overgrowth and inflammation in a human gut-on-a-chip,” Proc. Natl. Acad. Sci. U.S.A., vol. 113, no. 1, pp. E7–E15, Jan. 2016.
- Emulate Inc. (n.d.). [Online].
- J. W. Kotula, S. J. Kerns, L. A. Shaket, L. Siraj, J. J. Collins, J. C. Way, and P. A. Silver, “Programmable bacteria detect and record an environmental signal in the mammalian gut,” Proc. Natl. Acad. Sci. U.S.A, vol. 111, no. 13, pp. 4838–4843, Apr. 2014.



