As antibacterial resistance increases—now thought responsible for an estimated 5 million deaths worldwide each year [1]—scientists are investigating two distinct approaches to combating this deadly problem: Develop new antibiotics that defy pathogen resistance or design methods that hone antibiotic selection and fight individual infections better.
New antibiotic for growing foe
In May 2023, researchers announced that artificial intelligence (AI) had helped them discover a compound to fight an especially difficult-to-kill, hospital-acquired bacterial infection. A recent study [2] put the estimated attributable mortality rate of Acinetobacter baumannii infection at between 8.4% and 36.5%.
“This bacterium, Acinetobacter baumannii, likes to hang out on medical equipment, doorknobs, and other surfaces, and the reason it’s becoming such a challenge is that it has the unique propensity to acquire extracellular DNA—which in hospitals often encodes antibiotic resistance genes—and keep it,” said Jonathan Stokes (Figure 1), Ph.D., assistant professor of biochemistry and biomedical sciences at McMaster University, and coauthor of the Nature Chemical Biology article detailing the finding. “In addition, Acinetobacter is really virulent and causes lung, blood, wound, and other infections that can be fatal.”

Figure 1. Jonathan Stokes of McMaster University (shown here), and his research group have identified a new antibacterial compound to treat Acinetobacter baumannii, a highly resistant bacterium that causes dangerous hospital-acquired infections. (Photo courtesy of Matt Clarke/McMaster University.)
The research group’s AI algorithm grew from 2019 work at the Massachusetts Institute of Technology (MIT), which included developing an architecture—known as a message-passing neural network—to predict the physicochemical properties of molecules [3]. In 2020, James Collins, Ph.D., MIT Termeer Professor of Medical Engineering and Science and a professor of biological engineering, and Stokes, who was then a post-doctoral at MIT, got involved and tried using the AI algorithm to predict molecular properties for antibiotics against lab-strain Escherichia coli. It worked [4].
Once Stokes got a faculty position at McMaster, he decided to investigate the truly problematic pathogen Acinetobacter baumannii. To do so, he and his research group screened 7,500 chemicals to create a training dataset that identified both those compounds that fought Acinetobacter, and those that had no effect on other bacteria (Figure 2). They then trained the model on the training dataset, so that it was capable of looking at compounds it had not seen before and predicting which of those could kill Acinetobacter baumannii.

Figure 2. Graduate student Gary Liu was part of the Stokes group that screened 7,500 chemicals to train an AI model to wade through compounds and predict which could kill A. baumannii. (Photo courtesy of Matt Clarke/McMaster University.)
When Stokes’ group tested the model’s predictions in the laboratory, one structurally distinct compound rose to the top as being both potent and highly targeted against Acinetobacter baumannii. “The model wasn’t trained to pick out narrow-spectrum antibiotics—that was just an interesting observation from the wet lab—but that’s when we thought, ‘We found something really interesting here,’” Stokes recalled. Because of its targeted nature, the new antibiotic doesn’t kill the good bacteria with the bad, so the patient has better health outcomes, and “it doesn’t impose a universal selective pressure, so it decreases the horizontal dissemination of resistance,” he explained.
Since that initial discovery, the group has tested the new molecule, which they have named abaucin (Figure 3), on wounds in mice and found that it effectively suppresses Acinetobacter baumannii [5]. “We didn’t expect sterilization. Rather, we expected that it would prevent this bacterium from growing and turning into a really nasty infection, and that is what we observed in mice,” Stokes said. The researchers have also conducted genetics, omics, and genetic engineering experiments to determine abaucin’s mechanism of action and believe it inhibits an essential bacterial pathway known as lipoprotein trafficking.

Figure 3. Liu and graduate student Denise Catacutan (shown here) were two of the coauthors on the paper announcing the discovery of the new antibacterial compound, named abaucin, and showing its bacteria-suppressing capabilities on wounds in mice. (Photo courtesy of Matt Clarke/McMaster University.)
While excited about the prospects of abaucin, Stokes noted that getting it from the laboratory to the clinic will require a lot of work. “We’ve done steps one through five of a multihundred-step process and are now focusing on things like optimizing the structure of the chemical to improve its potency against Acinetobacter, as well as investigating medicinal properties such as pharmacokinetics, pharmacodynamics, solubility, and more,” he said. “At this point, we’re cautiously optimistic and truly excited about the prospects of abaucin since it has a novel mechanism of action—a really interesting narrow-spectrum activity—and there is a strong clinical need.”
Matching antibiotic to pathogen
One researcher looking to improve antibiotic selection is George Tetz, M.D., Ph.D., of the nonprofit Human Microbiology Institute, and CEO of the startup TGV-Dx, both of New York, New York. The focus of his research group and the company is on developing more accurate and faster methods for doctors to select the most effective antibiotics for their patients, particularly those who are struggling with infections caused by hard-to-treat, multidrug-resistant pathogens. This includes patients who have cystic fibrosis with notorious and challenging Pseudomonas aeruginosa infections, ventilator-associated pneumonia, and relapsing and recurrent urinary tract infections (UTIs).
The researchers have now developed a diagnostic test, called AtbFinder, which is designed to address “flaws” in current methods of antibiotic selection, Tetz said. One of these “flaws” is speed. Conventional antimicrobial susceptibility testing (AST) methods take 48–96 hours to yield results, but since patients often cannot wait that long, physicians make educated guesses, or so-called empiric selections, of which antibiotics to prescribe. “Unfortunately, among patients who have hard-to-treat bacterial infections or who have an infection caused by a multi-drug-resistant bug, these empirically selected antibiotics frequently fail to be effective,” he said.
Another issue is accuracy. “Even after the AST results come from the lab, in up to 10% of cases, they fail to select the right antibiotic. Why? Because AST centers on the response to antibiotics only of the lead bacterial pathogen and ignores the critical roles supporting bacteria may play in the antibiotic response of the lead bacterial pathogen,” Tetz said. Those roles include secreting bacteria-protecting agents, participating in the creation of defensive biofilms, and modulating the expression of antibiotic-resistant genes in the lead bacterium. AtbFinder is designed to fill both gaps by selecting antibiotics based on the response of the entire microbial community at the site of the infection and by providing rapid results.
AtbFinder uses 48-well plates filled with a unique host-mimicking growth medium called TGV agar, which recaptured the entire polymicrobial community from the clinical biosamples in AtbFinder, Tetz explained, noting that each well also is supplemented with one to several antibiotics at concentrations achievable at the particular site of infection. “With AtbFinder,” he related, “you plate the biosamples directly on the surface of the wells of TGV agar, incubate the plates, and within just 4 hours, you can see with the naked eye the presence or absence of microbial growth.” By running samples on four plates, a user can quickly see the effectiveness of nearly 200 antibiotics and combinations, he added (Figure 4).
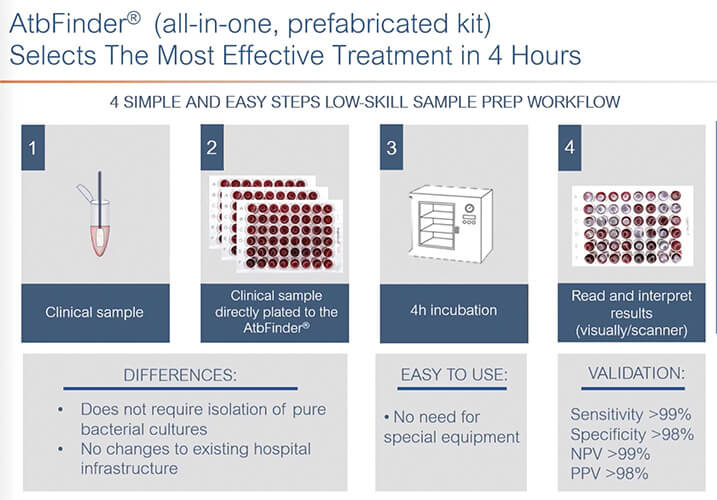
Figure 4. Researchers at TGV-Dx have developed a diagnostic test, called AtbFinder, that selects antibiotics based on the response of the entire microbial community at the site of the infection, rather than the response of only the lead bacterial pathogen. This is important, according to CEO George Tetz, because supporting bacteria can affect the overall antibacterial response. (Image courtesy of TGV-Dx.)
They have tested AtbFinder on 35 individuals who have cystic fibrosis and chronic Pseudomonas aeruginosa infections that are not resolved with AST-determined antibiotics [6]. “We found that AtbFinder was able to select antibiotics that resulted in the eradication of multi-drug resistance in P. aeruginosa in more than 81% of patients,” he said. The study also reported marked improvement in lung function, going from 44.2% forced expiratory volume (air exhaled in a forced breath) to 65.1%, and a significant reduction in chronic inflammatory markers across patient populations. In addition, the research group looked at hospitalizations due to pulmonary exacerbations in the two years prior to the study when the 35 patients were taking AST-selected antibiotics, and the two years they were taking AtbFinder antibiotics. Under AST-selected antibiotics, the patients had a total of 85 hospitalizations in two years, and under AtbFinder-selected antibiotics, the patients had two.
The researchers found similar results with recurrent UTIs. In a case study [7] of a patient who had a chronic UTI that continued despite six courses of AST-selected antibiotics over 24 months, “just one course of AtbFinder-selected antibiotics (yielded) a total disappearance of symptoms, and after a second course, there was a complete eradication of the infection. Currently, the patient has been free of UTI for more than a year,” Tetz reported.
TGV-Dx is now setting up a proprietary clear lab to produce the AtbFinder well plates so it can get them to physicians as a laboratory-developed test and hopefully help patients with infections not resolved with AST-selected antibiotics. In this way, Tetz remarked, “We believe we can make a difference.”
New test and new antibiotic
A group in California also hopes to improve the traditional AST methods, and their tactic involves replacing the standard nutrient agar with body-mimicking cell culture, according to project lead Michael Mahan, Ph.D., of the Department of Molecular, Cellular, and Developmental Biology, University of California, Santa Barbara (UCSB) (Figure 5). “The standard growth medium is super-cheap, widely available, and many pathogens will grow on it, but it doesn’t reflect the body. And bacteria behave and respond differently in the body than they do in nutrient agar.”

Figure 5. Michael Mahan at the Department of Molecular, Cellular, and Developmental Biology, University of California, Santa Barbara, led projects that developed both a new antibiotic and a new antibiotic test. (Photo courtesy of Michael Mahan.)
For this project, Mahan and his group compared results from the traditional AST assay and one that replaced the standard medium with cell culture and followed up by validating their cell-culture results in mouse models [8], [9]. In their first experiments with more than 1,300 different combinations of bacteria and antibiotics, their altered AST assay recommended a completely different antibiotic in one out of ten tests. “This means that 10% of the time, we might be prescribing the wrong drug,” he contended.
Most recently, the researchers tested antibiotics recommended by the altered AST assay in more than 25 different animal models of infection [10]. This exhaustive study confirmed their earlier results: AST is much more accurate with cell culture (Figure 6), he said.
Figure 6. Mahan’s group tweaked the currently used antibiotic susceptibility test and developed one they have shown to be more accurate in selecting effective antibiotics. Participating institutions in this project included Santa Barbara Cottage Hospital, Santa Barbara, CA, USA, and the University of Sydney’s School of Veterinary Science, Sydney, NSW, Australia. (Image courtesy of Ryan Allen and Peter Allen, Second Bay Studios.)
The study also showed that many AST-discounted antibiotics are actually highly effective against a variety of bacterial infections, including superbugs. This is nothing new to physicians, Mahan remarked. “For instance, the antibiotic azithromycin, also known as Z-pak, is the second-most-prescribed drug in the United States, but it’s never recommended for methicillin-resistant Staphylococcus aureus (MRSA) infections because it fails the standard AST results. Yet, doctors will often go ‘off-menu’ and prescribe it anyway, because they have seen that it can work.”
At the same time, Mahan’s group was also working on a new antibiotic that actually started out as a potential means to develop a long-lasting cell phone battery for U.S. Army troops. For the Army project, UCSB chemist Guillermo Bazan and his group created simple chemical compounds, called conjugated oligoelectrolytes (COEs), that could access the energy bacteria use to reproduce, and transform it into a power source. The bacteria did not survive well, which led to the researchers thinking about adapting the molecules to become antibiotics.
Bazan approached Mahan about testing some of the COEs they had created. As is typical for the Mahan lab’s approach, the researchers did their initial experiments in animal models. “We asked two things: Is it toxic? And, can it clear untreatable infections?” Of the 17 COEs they first investigated, only one was nontoxic. “And not only wasn’t it toxic, but it could kill every gram-negative and -positive pathogen we tested, and we have a lot in the lab. That includes MRSA and carbapenem-resistant Klebsiella pneumonia, which is resistant to nearly every drug and probably the most feared pathogen in the United States,” he said (Figure 7).

Figure 7. Mahan’s group also developed a highly effective and seemingly resistance-proof antibiotic. Participating research groups and institutions included the UCSB Institute for Collaborative Biotechnologies, the Infectious and Inflammatory Diseases Research Center in La Jolla, CA, USA, Santa Barbara Cottage Hospital, and the University of Sydney’s School of Veterinary Science. (Image courtesy Ryan Allen and Peter Allen, Second Bay Studios.)
In collaboration with microbiologist Andrei Osterman and his group at the Sanford Burnham Prebys Medical Discovery Institute in La Jolla, California, they found another unusual benefit of the killer antibiotic: Bacterial pathogens were unable to form resistance to it [11]. “That is just unheard of, and it is what makes this really special,” Mahan said, noting that the antibiotic’s “irresistibility” apparently stems from its multiple modes of action. “It affects many functions, such as a bacterium’s energy metabolism and just about anything to do with the membrane, so a bacterium would have to have mutations in all the affected pathways to override it.”
Despite all these positive aspects, the new antibiotic will not be in clinical use anytime soon. “These COEs are synthetic molecules that were made for an entirely different purpose and have never before been seen on planet Earth, so while our short-term study showed it wasn’t toxic to mice, we need to investigate what happens over a longer period and at different doses,” Mahan said. “And we’re working on this feverishly.”
His fervor stems from the ongoing threat antibiotic resistance currently poses. Whether the answer lies in new antibiotics, better testing to find optimal antibiotics or a combination of the two, the time has come to focus on this growing global health crisis, he said. “We already have drug-resistant staph, strep, and tuberculosis infections that do not respond to existing medications. People are dying from urinary tract infections. This is going to happen more and more. So, we need to get on this quickly, and not in a couple of years—now.”
He added, “I think the academic world and the biotech world can solve the research questions, but the implementation of the solution is going to be carried out by pharmaceutical companies. To make that happen, society is going to have to make some decisions, and soon.”
References
- Antimicrobial Resistance Collaborators, “Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis,” Lancet, vol. 399, no. 10325, pp. 629–655, Feb. 2022, doi: 10.1016/S0140-6736(21)02724-0.
- H. J. Appaneal et al., “Treatment, clinical outcomes, and predictors of mortality among a national cohort of admitted patients with Acinetobacter baumannii infection,” Antimicrobial Agents Chemotherapy, vol. 66, no. 3, Mar. 2022, Art. no. e01975.
- K. Yang et al., “Analyzing learned molecular representations for property prediction,” J. Chem. Inf. Model., vol. 59, no. 12, pp. 5304–5305, Dec. 2019.
- J. M. Stokes et al., “A deep learning approach to antibiotic discovery,” Cell, vol. 180, no. 4, pp. 688–702, Feb. 2020.
- G. Liu et al., “Deep learning-guided discovery of an antibiotic targeting Acinetobacter baumannii,” Nature Chem. Biol., May 2023, doi: 10.1038/s41589-023-01349-8.
- G. Tetz et al., “AtbFinder diagnostic test system improves optimal selection of antibiotic therapy in persons with cystic fibrosis,” J. Clin. Microbiol., vol. 61, no. 1, Jan. 2023, Art. no. e01558.
- G. V. Tetz et al., “Treatment of chronic relapsing urinary tract infection with antibiotics selected by AtbFinder,” Urol. Case Rep., vol. 46, Jan. 2023, Art. no. 102312.
- J. Z. Kubicek-Sutherland et al., “Host-dependent induction of transient antibiotic resistance: A prelude to treatment failure,” eBioMedicine, vol. 2, no. 9, pp. 1169–1178, Sep. 2015.
- S. C. Ersoy et al., “Correcting a fundamental flaw in the paradigm for antimicrobial susceptibility testing,” eBioMedicine, vol. 20, pp. 173–181, Jun. 2017.
- D. M. Heithoff et al., “Re-evaluation of FDA-approved antibiotics with increased diagnostic accuracy for assessment of antimicrobial resistance,” Cell Rep. Med., vol. 4, no. 5, May 2023, Art. no. 101023, doi: 10.1016/j.xcrm.2023.101023.
- D. M. Heithoff et al., “A broad-spectrum synthetic antibiotic that does not evoke bacterial resistance,” eBioMedicine, vol. 89, Mar. 2023, Art. no. 104461, doi: 10.1016/j.ebiom.2023.104461.