Artificial intelligence (AI) and machine learning (ML) have influenced medicine in myriad ways, and medical imaging is at the forefront of technological transformation. Recent advances in AI/ML fields have made an impact on imaging and image analysis across the board, from microscopy to radiology. AI has been an active field of research since the 1950s; however, for most of this period, algorithms achieved subhuman performance and were not broadly adopted in medicine. Recent enhancements for computational hardware is enabling researchers to revisit old AI algorithms and experiment with new mathematical ideas. Researchers are applying these methods to a broad array of medical technologies, ranging from microscopic image analysis to tomographic image reconstruction and diagnostic planning.
Despite promising academic research in which algorithms are beginning to outperform humans, healthcare delivery in the most advanced nations of the world still has minimal AI involvement [1]. The hopes and tribulations brought by AI in medicine have been a matter of discussion and debate. On one hand, AI promises to deliver improved diagnosis, enable early detection, and uncover novel personalized treatments; on the other, it may threaten jobs for millions and might disrupt the medicolegal ecosystem. Many countries have initiated independent policymaking bodies to study the broad socioeconomic effects of AI and generate national plans for strategic AI development [2].
In this discussion, we focus primarily on emerging directions for AI in medical imaging and related technical challenges. We also discuss AI’s broader impact and consider the views of young researchers and medical professionals, who represent key stakeholders within the new ecosystem.
Recent Advances in AI
While the fields of signal processing, optimization, and adaptive systems have seen extensive research during the last 70 years, recent advances in computational power have led to new techniques and significant improvements in image recognition, speech processing, and a variety of other areas that traditionally have been considered part of AI. For example, while convolutional neural networks (CNNs) appeared in the literature at least as far back as 1990, these structures were not considered widespread until the publication of a 2012 study on ImageNet classification [3]. Major trends in AI research include deep reinforcement learning and generative adversarial networks. Figure 1 depicts some recent advances in AI platforms and related medical innovations that employ these technologies.
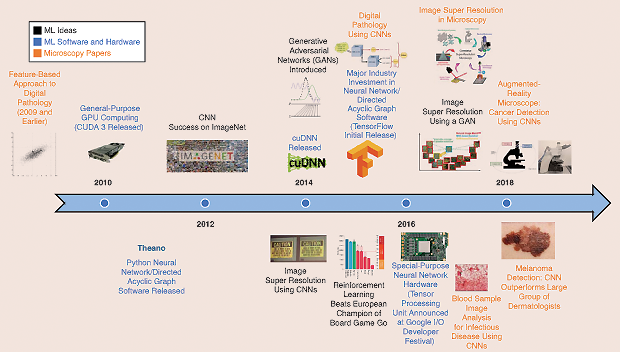
A common theme underpinning recent advances is the idea of deep feature representations. In the past, data in a high-dimensional space were represented using a lower-dimensional set of features. These features were typically either chosen by a human or computed using techniques such as singular value decomposition. Rather than computing a single set of features, a deep feature representation learns many layers of features, where each layer (usually) decreases the dimensionality of the data. These features may be learned in a supervised or unsupervised manner, depending on the mathematical methods used. Eventually, the final feature layer will be much smaller in dimension than the original input and may be employed in conjunction with traditional estimation or classification techniques. The ability to learn deep feature representations in practice has been enabled by special-purpose computer hardware, such as graphics processing units (GPUs), and custom chips, such as Google’s Tensor Processing Unit.
AI in Pathology
Microscopy Image Formation: Super Resolution
In the context of microscopy, enhanced image resolution allows one to view digital slides in more detail or to scan whole-slide images using fewer pictures, thereby reducing scan time. While standard techniques, such as linear and bicubic interpolation, increase the number of pixels in an image, they do not necessarily increase image detail. However, because interpolation is essentially a prediction problem, deep learning techniques, such as CNNs and generative methods, have been applied to space, resulting in a set of super-resolution algorithms for increasing image resolution. These new methods are now becoming commonplace in the microscopy community, enabling higher-resolution scans and increased scanner throughput. In particular, super-resolution techniques have decreased the light microscopy minimum linewidth from approximately 300 nm to approximately 10 nm within the last decade [4]. When scanning a section of a slide, typical whole-slide imaging devices take photographs on a regular grid and stitch together the final image. Using super-resolution techniques, the image grid can be up to two orders of magnitude more sparse [5], thereby reducing the required number of photographs and increasing slide scanning throughput.
Microscopy Image Analysis
Image Segmentation
Segmentation has been a long-standing problem in the image-processing community, with standard techniques including morphological operators, thresholding, and the watershed algorithm. Deep learning methods are now employed for image segmentation, thereby enhancing the performance of any algorithm that uses image segmentation as a preprocessing step. For example, in electron microscopy, CNNs have been employed for segmentation for three-dimensional brain imaging [6]. Generative models have been employed for cellular boundary detection [7]. Now that deep learning software is being incorporated in widely used image-processing libraries, such as OpenCV, one can expect that a broad array of algorithms will be improved by enhanced image segmentation tools.
Digital Pathology
One application of image classification is automated location and scoring of tumors for digital pathology. However, standard CNN techniques are designed to take a fixed-size image patch as an input and produce a single class label. In digital pathology, one would like a class label for each image pixel rather than for the entire image. Fully convolutional neural networks, which predict a heatmap rather than a single class label, are ideal for these applications. This approach is currently employed for real-time cancer detection as part of an augmented-reality microscope [8]. Prior approaches to cancer detection typically use patch-based methods, wherein the entire input image patch corresponds to a single class and the scoring of a whole slide is accomplished by scoring many individual patches. Using heatmap methods substantially increases neural network training and inference performance, which is important for the large data sets necessary to create robust classification systems.
[accordion title=”How AI Is Optimizing the Detection and Management of Prostate Cancer”]
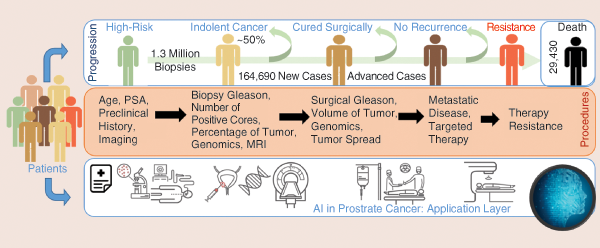
Annually, approximately 20 million men are prostate-specific-antigen screened, and 1.3 million undergo an invasive biopsy to diagnose roughly 200,000 new cases, 50% of which end up being indolent. Approximately 30,000 men die of prostate cancer (PCa) yearly. Importantly, an estimated US$8 billion is spent on unnecessary biopsies. Thus, an integrative analysis and predictive model of prognosis is needed to help identify only lethal and aggressive forms of the disease.
Development toward achieving this goal has been fueled by tremendous advances in the field of genomics, imaging [especially magnetic resonance imaging (MRI)], and biomarker (such as exosomes and molecular imaging) research. The integrative power of artificial intelligence (AI) and machine learning holds immense promise in altering the identification of aggressive PCa (see Figure S1). One example is the use of genomics, AI, and extracellular vesicles, including exosomes and cell-free DNA from body fluids, to build a noninvasive PCa test [S1], [S2].
Another example is the use of AI in pathology, where several studies have demonstrated the machine-based identification of Gleason scores from pathology slides. Through the advancement of high-resolution MRI, a more detailed picture of the prostate is now possible. By integrating the features thus captured with genomics and gold-standard pathology, there have been efforts to predict aggressive PCa through noninvasive scans [S3], [S4].
The roles of diet, exercise, and lifestyle habits have remained a confounding factor. Identifying these environmental influences is highly challenging and requires laborious combing through mountains of unstructured clinical and nonclinical data. Efforts are underway to identify these using AI and natural language processing tools. Once available, these can be combined with the previously mentioned advancements to accurately identify patients who are in need of aggressive treatments.
Contributed by Kamlesh K. Yadav, M.Tech., Ph.D., assistant professor of urology at the Icahn School of Medicine at Mount Sinai, New York and principal scientist with Sema4 Genomics, Stamford, Connecticut. He has expertise in basic prostate oncology and therapeutics and is interested in combining these with genomics and AI for better disease diagnosis and prognosis.
References
[S1] J. McKiernan, M. J. Donovan, V. O’neill, S. Bentink, M. Noerholm, S. Belzer, J. Skog, M.W. Kattan, A. Partin, G. Andriole, and G. Brown, “A novel urine exosome gene expression assay to predict high-grade prostate cancer at initial biopsy,” JAMA Oncology, vol. 2, no. 7, pp. 882–889, 2016.
[S2] E. Hodara, D. Zainfeld, G. Morrison, A. Cunha, Y. Xu, J. D. Carpten, P. V. Danenberg, P. W. Dempsey, J. Usher, K. Danenberg, and F.Z. Bischoff, “Multi-parametric liquid biopsy analysis of circulating tumor cells (CTCs), cell-free DNA (cfDNA), and cell-free RNA (cfRNA) in metastatic castrate resistant prostate cancer (mCRPC),” J. Clinical Oncology, vol. 36, no. 6, Feb. 2018. doi: 10.1200/JCO.2018.36.6_suppl.274.
[S3] A. Salmasi, J. Said, A. W. Shindel, P. Khoshnoodi, E. R. Felker, A. E. Sisk, T. Grogan, D. McCullough, J. Bennett, H. Bailey, H. J. Lawrence, D. A. Elashoff, L. S. Marks, S. S. Raman, P. G. Febbo, and R. E. Reiter, “A 17-gene Genomic Prostate Score assay provides independent information on adverse pathology in the setting of combined mpMRI fusion-targeted and systematic prostate biopsy.” J. Urology, 2018. doi: 10.1016/j.juro.2018.03.004.
[S4] A. T. Beksac, S. Cumarasamy, U. Falagario, P. Xu, M. Takhar, M. Alshalalfa, A. Gupta, S. Prasad, A. Martini, H. Thulasidass, R. Rai, M. Berger, S. Hectors, J. Jordan, E. Davicioni, S. Nair, K. Haines, III, S. Lewis, A. Rastinehad, K. Yadav, I. Jayaratna, B. Taouli, and A. Tewari, “Multiparametric MRI features identify aggressive prostate cancer at the phenotypic and transcriptomic level,” J. Urology, 2018. doi: 10.1016/j.juro.2018.06.041.
[/accordion]
Infectious Diseases
Performing disease diagnosis on a blood sample is strongly preferable to using stained tissue samples, as is common in pathology. Deep learning methods are now used in numerous approaches; blood cultures can be interpreted using CNNs. For example, a bloodstream has been interpreted in a way similar to a communications system having a bit error rate [9], with the “interesting” cells occuring at approximately the same probability rate as erroneous bits. Further, many groups are analyzing gene expression data from blood samples using a variety of deep learning methods [10].
[accordion title=”AI and Clinicians: Not a Mutually Exclusive Zero-Sum Game”]
Recent bold, eye-catching headline predictions made by nonradiologists, e.g., “in a few years, radiology will disappear” and “stop training radiologists now,” are not only far from reality but also irresponsible and a disservice to the appropriate implementation and adoption of artificial intelligence (AI) technology to health care. It is highly likely and foreseeable that AI will enhance the quality and efficiency of the current clinical practice across many specialties and even render some activities in clinical practice obsolete.
While radiologists are all in agreement that AI will change and improve clinical radiology practice, AI technology in its current form is not ready for prime time. It will take a long while for many silos of machine learning to fully mature and be practiced in clinical radiology. Radiologists are pragmatic and willing to embrace new techniques that would enhance patient care and radiology as a medical specialty. Around the world, radiologists are inundated with clinical workloads and would welcome an AI solution to improve patient care and job satisfaction. However, we must go through many rounds of rigorous testing and evaluation to integrate AI technology into a long, complex network of activities involved in clinical radiology practice.
Producers and developers of AI technology should focus not only on high-hanging clinical applications such as radiomics but also on low-hanging practical solutions to improve many daily workflow-related clinical problems. Eventually, when AI plays a significant role in many practice activities, radiologists will gain enhanced efficiency and quality that could be translated into a reduction in the radiology workforce. Nevertheless, it is important to note that radiologists and AI are not mutually exclusive in a zero-sum game. As radiologists adopt AI technology to form a partnership, there will be a greater gain in the economic surplus as a whole that can be redistributed and shared with patients, providers, and payers to enhance the overall value in health care.
Contributed by Kyongtae Ty Bae, Ph.D., M.D., M.B.A., associate dean of clinical imaging research and professor of radiology and bioengineering at the University of Pittsburgh, Pennsylvania. He wears multiple hats as an engineer, radiologist, administrator, academician, and entrepreneur.
[/accordion]
AI in Radiology
Radiology involves a complex interplay of physics, engineering, and medicine and provides a rich playground for AI. Researchers have employed AI algorithms to solve problems ranging from image reconstruction to image analysis and computer-aided diagnosis.
Medical Imaging Engineering and Radiomics
The reconstruction of tomographic images (magnetic resonance, X-ray-computed tomography, etc.) is challenging due to sensor nonidealities and noise. These probabilistic distortions make the exact analytic inverse transform for image reconstruction difficult. Newer methods are adopting data-driven learning models to map the transformation between sensor and image spaces, instead of relying on complex signal processing of the acquired data and its associated parameter-tuning difficulties. Once the image is formed, the next challenge is to determine clinically relevant features. AI has traditionally been used most widely in image analysis of radiological data. As data sets grow, the number and efficacy of such analyses have steadily improved.
More recently, researchers have combined images with quantitative clinical outcomes, leading to the emergence and expansion of radiomics [11]. Moreover, the field of imaging genomics, where radiomics data and genomic information are combined, has gained momentum. Recent studies employing radiomics and imaging genomics have quantified heterogeneity and provided insights into disease transformation, progression, and drug resistance—providing new avenues for achieving personalized medicine.
The high-throughput, data-driven personalized care regime captures only one aspect of the promise of AI in health applications. As a technology that scales at low marginal cost, AI has the potential to deliver large-scale healthcare screening services to impoverished populations in resource-scarce communities. Such healthcare delivery opportunities have created considerable academic interest; however, clinical implementation will require the navigation of substantial political, financial, and regulatory hurdles.
Computer-Aided Diagnostics Systems for Radiology Practice
While the intellectual challenges associated with applying AI in health care are academically interesting, equally important is understanding how caregivers will react to the adoption of powerful new technologies that profoundly alter their profession. Computer-aided diagnosis tools can be sorted into three broad categories [12]:
- AI-replace (AI-R), aimed at substituting AI for the work of the radiologist
- AI-assist (AI-A), with the goal of helping the radiologist perform better
- AI-extend (AI-X), which uses AI to perform altogether new tasks not possible before.
AI-R systems can deliver results and outcome metrics without the assistance of trained readers and have shown great applicability not only at tertiary healthcare centers but also in far-flung regions where there are no radiologists (e.g., the BoneXpert software for automated measurement of bone age from a child’s hand X-ray). AI-A systems augment the capability of the reader (e.g., Veolity for lung cancer screening). Studies have reported a more than 40% improvement in the reading time of radiologists using such systems. More importantly, AI-A helps the intervening physician to interpret the data directly instead of always requiring the advice of a trained radiologist; good examples are found in ophthalmology and dermatology. In time, these systems may be upgraded to the AI-R category. Finally, AI-X systems add real value to the healthcare system by allowing us to detect ailments that cannot be read even by trained radiologists and enabling us to get more out of the captured data. Icomatrix for Multiple Sclerosis lesion detection and a newly developed method (by Google Inc.) for predicting cardiovascular risk factors from retinal fundus images [13] are good examples of this emerging class of software systems.
AI systems help us provide better diagnostic and treatment outcomes and can further enable the use of data acquired for one clinical reason to predict the risk of another condition.
Looking forward, AI-based systems will change the status quo between treating physicians and imaging specialists such as radiologists and pathologists. These new systems allow for direct assessment by the attending physician and will force the traditional imaging specialist to add more value to the diagnostic outcome, likely giving rise to new fields and professions. However, all studies unequivocally agree that, despite the disruption caused by AI technologies, medical imaging enhanced by ML will improve patient outcomes in an increasingly large and diverse global population (see “How AI Is Optimizing the Detection and Management of Prostate Cancer”).
[accordion title=”Fine-Tuning Deep Learning by Crowd Participation”]
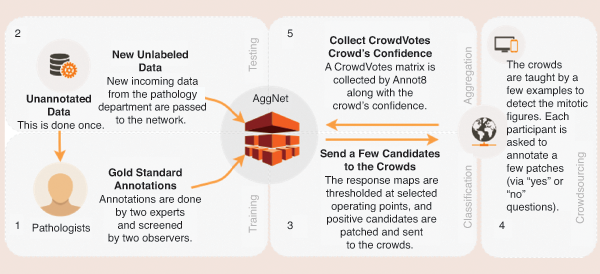
One of the major challenges currently facing researchers in applying deep learning (DL) models to medical image analysis is the limited amount of annotated data. Collecting such ground-truth annotations requires domain knowledge, cost, and time, making it infeasible for large-scale databases. Albarqouni et al. [S5] presented a novel concept for learning DL models from noisy annotations collected through crowdsourcing platforms (e.g., Amazon Mechanical Turk and Crowdflower) by introducing a robust aggregation layer to the convolutional neural networks (Figure S2). Their proposed method was validated on a publicly available database on breast cancer histology images, showing astonishing results of their robust aggregation method compared to the baseline of majority voting. In follow-up work, Albarqouni et al. [S6] introduced the novel concept of a translation from an image to a video game object for biomedical images. This technique allows medical images to be represented as star-shaped objects that can be easily embedded into a readily available game canvas. The proposed method reduces the necessity of domain knowledge for annotations. Exciting and promising results were reported compared to the conventional crowdsourcing platforms.
Contributed by Shadi Albarqouni, Ph.D., currently a senior research scientist and chair for computer-aided medical procedures at TU Munich. His research interests include semi-/weakly supervised deep learning (DL), domain adaptation, uncertainty, and explainability of DL models.
References
[S5] S. Albarqouni, C. Baur, F. Achilles, V. Belagiannis, S. Demirci, and N. Navab, “AggNet: Deep learning from crowds for mitosis detection in breast cancer histology images,” IEEE Trans. Med. Imag., vol. 35, no. 5, pp.1313–1321, 2016.
[S6] S. Albarqouni, S. Matl, M. Baust, N. Navab, and S. Demirci, “Playsourcing: A novel concept for knowledge creation in biomedical research,” in Deep Learning and Data Labeling for Medical Applications, G. Carneiro, D. Mateus, P. Loïc, A. Bradley, J. M. R. S. Tavares, V. Belagiannis, J. P. Papa, J. C. Nascimento, M. Loog, Z. Lu, J. S. Cardoso, and J. Cornebise, Eds. Cham, Switzerland: Springer, 2016, pp. 269–277.
[/accordion]
Clinical Translation of Medical AI Technologies
The first half of 2018 has already celebrated the coming to market of three medical AI technologies approved by the U.S. Food and Drug Administration (FDA)—all in medical imaging. In February, Viz.AI’s ContaCT application received approval for using AI to detect, from computed tomography images, large-vessel-occlusion acute ischemic strokes. In April, IDx-DR became the first approved medical device that combines a special camera and ML to detect moderate to severe diabetic retinopathy in adults. And in May, OsteoDetect, which analyzes two-dimensional X-ray images for signs of distal radius fracture, also received approval. These de novo approvals represent more than the successes of individual technologies or companies; they indicate that the dust has finally settled in the debates over what constitutes adequate evidence of safety and quality for medical AI technologies (at least for now). These breakthroughs in medical device regulation will set precedents and accelerate future developments.
Like that for many emerging technologies, the regulatory landscape of medical AI technologies had been uncharted. AI technologies differ from traditional medical devices in that they are predominantly software, can be changed easily, and have shorter innovation cycles. This meant that new ways of evaluating and validating the safety and efficacy of these technologies need to be developed. Researchers typically deem an algorithm successful if it can identify a particular condition or ailment from images as effectively and accurately as pathologists and radiologists can. However, many claim that AI diagnostics should undergo the same rigorous clinical trials as those required for most drugs [14]. Increasingly, AI technologies are taking the well-worn path of extended clinical trials. Current data (at ClinicalTrials.gov) show that 35 clinical trials involving AI are at different stages of fulfillment (17 completed and 18 ongoing). The FDA Digital Health Program’s innovative Precert pilot, which allows regulators to endorse the research team as opposed to the product, is also likely to offer new opportunities for rapid and agile development.
Based on the recent FDA approvals, we may infer that AI technologies are extending the frontiers in terms of how they impact clinical practice. Early AI technologies currently being marketed already show some major trends in design strategies. First, technologies can be designed to act alongside physicians and rely on humans as the fallback mode in case of error. The ContaCT application, for instance, is designed to function as a parallel option in regular clinical workflow. It relies on the action of physicians as a safety net for mitigating the risks of false positives and false negatives in detection. Second, speed is often a major value proposition of AI technologies—as is seen in many comparative clinical trials that have been completed. Last but not the least, the use case for AI can involve enhancing the medical acumen and diagnostic capability of individuals and transferring these skills (and the corresponding responsibility) from specialists to generalists or even to patients themselves. The IDx-DR, for example, is the first device approved for analysis of retinal images to detect more than mild diabetes without the involvement of a clinician. Thus, the device can be used by healthcare providers who may not normally be involved in eye care. The IDx-DR and similar devices, if successfully deployed, are likely to bring about profound changes in the structure and dynamics of health care.
In developing countries, the same systems can enable primary caregivers and rural health centers to deliver value-added services even in absence of a radiologist and/or pathologist. Additionally, the constant evolution and emergence of new imaging modalities with ever-growing dimensionalities pose a challenge to experts to constantly update their skills [15]. AI can pave the way for seamlessly integrating information from multiple modalities across the imaging spectrum—and enable specialists to deliver the benefits of breakthrough imaging technologies to patients. (See “Opinion from the Expert” for the views of Prof. Kyongtae Ty Bae.)
Overall, examples such as the success of the FDA’s De Novo pathway and Breakthrough Devices Program are likely to continue to encourage AI innovation by medical device and digital health companies. However, while completing clinical trials and gaining FDA approvals are akin to moving mountains, obtaining scientific backup for the technology and regulatory clearances is only part of winning the clinical translation battle. Acceptance of the technology by the clinical community, proof of value to the players, demonstration of added value, review of new considerations of risk-sharing schemes, and supporting the medical–legal structure are some of the further downstream challenges.
Future Directions: Where Will AI Take Medicine?
While integrating AI in healthcare delivery systems may seem straightforward, the reality of clinical implementation will be a long journey. However, this journey is worth pursuing because medical AI technologies will revolutionize medical care; these technologies will allow doctors to focus on patients and clinical decisions rather than tedious, repetitive tasks. Factors such as the decreasing cost of computational hard- ware, increased interest in precision medicine, the growing availability of medical data, and the public’s impatience with the rising costs of drugs and medical care will also contribute to further adoption of AI technologies in health care.
Despite the great promise shown in the ap- plication of deep learning techniques to both mi- croscopy and radiology, there are technological challenges that need to be addressed.
- Many algorithms still have trouble with generalization. This is particularly evident when an algorithm is trained on data from one hospital and then applied to data at an- other hospital. While there has been some work in this area, there is still a need either for more diverse data sets or for algorithms that generalize better with limited data.
- Standardized, benchmark data sets need to be developed for algorithm research. The creation of the CIFAR and ImageNet benchmark data sets has enabled enormous advances in the image-recognition community; however,similar data sets do not exist for microscopy. There have been disease-specific data sets, such as Camelyon and the ISBI Challenge, but there is no agreed-upon benchmark with a wide range of microscopy data. The genomics community has done a much better job of making data public; similar efforts for microscopy are necessary to develop next-generation AI algorithms.
- Presently, most microscopy data are stored on glass slides; they are not digitized due to the high cost and low throughput of whole- slide image scanners. The development of low-cost, high-throughput scanners would assist in creating the necessary benchmark data sets.
- Data protection laws need to be internationally standardized to allow for medical research. For example, radiological data are governed by nonuniform data protection regimes of individual nations and territories. In particular, radiological data frequently cannot physically leave certain geographic locations due to legal restrictions. This makes it impossible for most researchers— especially computer scientists—to compile enough data for AI algorithm training. Therefore, many researchers are training separate models for each local set of data, leading to reduced algorithm performance and higher development costs. Building a global data set available to all researchers would dramatically increase the speed of medical AI development.
The potential of medical AI technology is well known. When asked, most clinicians can rattle off the list of potential benefits, including improving the speed, efficacy, and precision of medical treatments while reducing costs. Yet, interestingly, the technological innovators are more cautious. Instead of going after true intelligence and the goal of replacing physicians (although some still hold on to this vision), the focus of AI has largely moved from higher cognitive skills to classification. Furthermore, state-of-the-art development in AI is quite fragmented, in that each algorithm is designed to perform a specific task in a specific subspecialty of medicine—and, even then, only a few can outperform humans [16].
The emergence of AI in medicine and the positive changes it fosters can well outweigh the potential negative changes it threatens to bring. This current wave of AI technologies has been largely defined by open-software platforms and crowdsourcing (see “Fine-Tuning Deep Learning by Crowd Participation”). Open technologies enable remote diagnostic systems in impoverished communities [17], which represents only a small fraction of the benefits that AI offers the medical profession. As with all technological changes, the mechanics of AI itself merely open up a new set of opportunities; how these technologies are employed will be decided by humans, not machines.
References
- A. L. Fogel and J. C. Kvedar, “Artificial intelligence powers digital medicine,” npj Digital Med., vol. 1, no. 1, pp. 5, 2018.
- C. Cath, S. Wachter, B. Mittelstadt, M. Taddeo, and L. Floridi, “Artificial intelligence and the ‘good society’: The US, EU, and U.K. approach,” Sci. Eng. Ethics, vol. 24, no. 2, pp. 505–528, 2018.
- A. Krizhevsky, I. Sutskever, and G. E. Hinton, “ImageNet classification with deep convolutional neural networks,” in Proc. Advances Neural Information Processing Systems, 2012, pp. 1097–1105.
- M. Hauser, M. Wojcik, D. Kim, M. Mahmoudi, W. Li, and K. Xu, “Correlative super-resolution microscopy: New dimensions and new opportunities,” Chem. Rev., vol. 117, no. 11, pp. 7428–7456, 2017.
- W. Ouyang, A. Aristov, M. Lelek, X. Hao, and C. Zimmer, “Deep learning massively accelerates super-resolution localization microscopy,” Nature Biotechnol., vol. 36, no. 5, 2018. doi: 10.1038/nbt.4106.
- D. Ciresan, A. Giusti, L. M. Gambardella, and J. Schmidhuber, “Deep neural networks segment neuronal membranes in electron microscopy images,” in Proc. Advances Neural Information Processing Systems, 2012, pp. 2843–2851.
- M. Pachitariu, A. M. Packer, N. Pettit, H. Dalgleish, M. Hausser, and M. Sahani, “Extracting regions of interest from biological images with convolutional sparse block coding,” in Proc. Advances Neural Information Processing Systems, 2013, pp. 1745–1753.
- P.-H. Chen, K. Gadepalli, R. MacDonald, Y. Liu, K. Nagpal, T. Kohlberger, G. S. Corrado, J. D. Hipp, and M. C. Stumpe, “An augmented reality microscope for real-time automated detection of cancer,” in Proc. Annu. Meeting American Association Cancer Research, 2018.
- A. Mahjoubfar, C. Lifan Chen, and B. Jalali, Artificial Intelligence in Label-Free Microscopy. Berlin: Springer-Verlag, 2017.
- R. Fakoor, F. Ladhak, A. Nazi, and M. Huber, “Using deep learning to enhance cancer diagnosis and classification,” in Proc. Int. Conf. Machine Learning, vol. 28, 2013.
- A. Hosny, C. Parmar, J. Quackenbush, L. H. Schwartz, and H. J. W. L. Aerts, “Artificial intelligence in radiology,” Nature Rev. Cancer, vol. 18, no. 8, pp. 500–510, Aug. 2018.
- B. van Ginneken, “Artificial intelligence and radiology: A perfect match. Radiology and radiologists: A painful divorce?” presented at the European Congr. Radiology, 2018.
- R. Poplin, A. V. Varadarajan, K. Blumer, Y. Liu, M. V. McConnell, G. S. Corrado, L. Peng, and D. R. Webster, “Prediction of cardiovascular risk factors from retinal fundus photographs via deep learning,” Nature Biomedical Eng., vol. 2, no. 3, p. 158, Mar. 2018.
- Editorial, “AI diagnostics need attention,” Nature, vol. 555, no. 285, 2018. doi: 10.1038/d41586-018-03067-x.
- S. Mandal, X. L. Dean-Ben, N. C. Burton, and D. Razansky, “Extending biological imaging to the fifth dimension: Evolution of volumetric small animal multispectral optoacoustic tomography,” IEEE Pulse, vol. 6, no. 3, pp. 47–53, 2015.
- B. E. Bejnordi, M. Veta, P. J. Van Diest, B. Van Ginneken, N. Karssemeijer, G. Litjens, J. A. W. M. Van Der Laak, M. Hermsen, Q.F. Manson, M. Balkenhol, and O. Geessink, “Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer,” J. Amer. Med. Assoc., vol. 318, no. 22, pp. 2199–2210, 2017.
- S. Mandal, “Frugal innovations for global health—Perspectives for students,” IEEE Pulse, vol. 5, no. 1, pp. 11–13, 2014.



