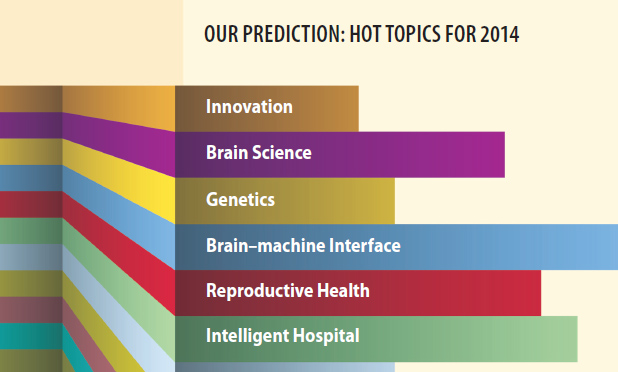
When it comes to predicting the future, everyone has their own approach. Weather forecasters track changing pressure systems, economists study the markets, and doctors wrestle with patient risk factors. And here at IEEE Pulse? Over the last few months, we’ve talked to experts in the field, asked IEEE Engineering in Medicine and Biology Society members for input, and scanned through the past year’s research breakthroughs to identify what we think might be the hottest biomedical engineering areas to watch in 2014.
The results? Some are obvious—like everything and anything to do with the brain. And of course, nanomedicine will continue what’s been a clear upward trajectory, while a brand-new genetic editing tool that took the world by storm earlier last year will settle into laboratories worldwide on its way to transforming basic genetic work. However, there are also a few areas we found that might not be at the front of everyone’s mind—fields such as diabetes, reproductive health, and health care delivery that have been undergoing evolutions of their own and are now standing on the verge of transformation.
We know it is not exhaustive—in a field this active and innovative, how could it be? And only time will tell how well we’ve really called our bets. But if we’re right, to paraphrase a source’s description of the coming brain–machine interface (BMI) revolution, it’s going to be like science fiction rewritten—into reality.
Consider this your field guide to the near future. And it starts in the brain.
The Year of the Brain
There has never been a better time to be in brain research than the present. Money is pouring in: there is the €1 billion Human Brain Project in the European Union (EU), the U.S. BRAIN initiative that plans to start with US$100 million just for 2014, and sprawling private ventures such as GlaxoSmithKline’s electroceutical program, which plans to invest US$50 million into neurotech startups and support brain circuits research in up to 20 laboratories worldwide. Meanwhile, technology is pushing the envelope of reality and opening the brain to unprecedented viewing—consider for example: Stanford University’s CLARITY technique that turns entire brains almost crystal-clear for network mapping; stem-cell-grown minibrains (cerebral organoids) from the Institute of Molecular Biotechnology in Vienna; nanowire probes and tissue matrices for in-depth sensing.
The two biggest stimulants here are the governmental initiatives, both of which will spill heavily into numerous international collaborations. The Human Brain Project kicked off last January when the European Commission awarded its prize to Henry Markram of the École Polytechnique Fédérale de Lausanne (EPFL), Switzerland, and his goal of recreating a human brain in a supercomputer (“A CERN for the brain,” the EPFL has called it). BRAIN got its start in April 2013, with President Obama’s proposed aim of mapping the human brain in action.
It’s still in the early days, of course—both initiatives are only really getting underway this year, and many details are still to come. But a few trends are already emerging. The EU project, for instance, will have a heavy emphasis on neuroinformatics and computation. The U.S. project will orient around technological advancement in neuron and circuit manipulation, recording, and imaging, not to mention analyses that can handle the inevitable reams of numbers. Engineering of all kinds—neuro-, nano-, electrical—will be crucial to this brain exploration. So too will be a trend toward basic knowledge. “Electrode development, recording amplifiers, and decoding algorithms—these are engineering approaches, but none of them make sense unless you’re doing basic science first,” says Andrew Schwartz, a neurophysiologist at the University of Pittsburgh. Indeed, Schwartz says, we actually know remarkably little about the brain and how it makes usable information. Filling in those blanks will become increasingly essential in understanding what results from the arriving new tools and research agendas.
According to Bin He, distinguished McKnight University professor of biomedical engineering and director of the Center for Neuroengineering at the University of Minnesota, “Understanding how the brain works is one of the most exciting frontiers in science and will have a tremendous impact on our society, considering that brain disorders cost hundreds of billions of dollars annually.” Dr. He also chaired a recent National Science Foundation workshop that identified types of brain mapping that will be essential challenges for the U.S. initiative.
Indeed, as University of California (UC) San Diego neuroengineer Todd Coleman phrases it, “This space is definitely going to explode.”
The Brain-Machine Interface
Man Melds with Machine
When it comes to the brain specifically, there is one area in particular that’s on everybody’s mind: BMI. It has been a long time coming, just out of reach for decades. But now? Thought-controlled robotic arms have delivered chocolate to quadriplegics, bionic legs have stepped up Chicago’s Willis Tower, and miniature helicopters have flown under the mental guidance of their creators. “There are so many advances [in the BMI area] that are taking place,” Coleman says. “It’s literally science fiction rewriting itself.”

Spurred by the global brain initiatives and by the needs of wounded veterans, research here is advancing quickly at all levels—smaller sensors, smarter signal processing, and basic neuroscience—but there are a few core trends that will still stand out from the crowd this year:
- Non-Invasive Interfaces: Much of the most accurate BMI work so far has come from experiments that interface sensors directly with a patient’s brain, rather than through the scalp’s skin and muscle. But if such mind-controlled prostheses are ever going to get close to mainstream, they can’t be done through a hole in the head—and those in the field know it. According to He, a wave of work into better noninvasive techniques is gaining steam worldwide, both in terms of the number of teams working on the problem and the progress they have been able to make. In fact, last year, He’s own laboratory was able to fly a minihelicopter in multiple directions—with a degree of accuracy comparable to that achieved in other work done with in-brain probes, using EEG signals acquired from a 64-electrode cap. Meanwhile, researchers like Coleman, in collaboration with John Rogers at the University of Illinois at Urbana–Champaign, have begun to move away from the electrode cap entirely by designing paper-thin electrode arrays that stick like temporary tattoos capable of detecting neural signals. “In the next year or few years, there will be big advancements in this particular area of science to advance mind-controlled medical devices using noninvasive sensors,” says He.
- Bidirectional Neuromodulation: Doctors have placed electrodes in brains for years to alleviate patients’ symptoms of dystonia and Parkinson’s tremors. But lately, researchers are taking that concept a step further. Medtronic has developed a new sensor aimed at looping that traditional one-way flow of information, creating devices that not only stimulate but also record brain activity in real time. That sensor–recorder is already approved for investigational use in the United States and has been implanted in patients with Parkinson’s disease. Even more daringly, researchers at the Lawrence Livermore National Laboratory in California have teamed up with Emory psychiatrist Helen Mayberg to create electrochemical neuroimplants that could not only record and stimulate electrically but also sample brain chemistry and release drugs directly to, for example, circuits driving depression. “That opens up a whole new class of science,” says Lawrence Livermore scientist Sat Pannu. “You can now start understanding how the brain operates. We all look at our brains as a series of neurons that are all interconnected electrically—but all those interconnections actually happen chemically.”
- Restoring Sensation: For 40 years, cochlear implants have dominated the sensory prosthetic field, but the field is now evolving fast. The artificial retina, crafted with breakthrough thin-film electrode array technology, has hit markets after winning approval in Europe three years ago and in the United States last year. Now the Lawrence Livermore researchers behind that technology are trying to translate its thin-film arrays into next-generation cochlear implants, which would effectively increase the number of frequency channels a patient hears from a couple dozen to hundreds or thousands—potentially restoring hearing loss to the point of restoring the full range of musical experience. Beyond sight and hearing, work on perceptions such as touch is also gaining speed. Silvestro Micera, with the Center for Neuroprosthetics at the EPFL, is developing a bidirectional bionic hand harnessed to the peripheral nerves that not only moves on command but sends touch signals back to the brain as well. Early work in humans proved the technology successful at improving sensory feedback, and the arm is now headed toward bigger clinical trials in Italy. Similar work is underway elsewhere, including research by Todd Kuiken at the Rehabilitation Institute of Chicago, Illinois, where he’s expanding his pioneering idea of targeted sensory innervation—rerouting the nerves from a missing limb back to muscles upstream—to likewise improve mental control of bionic arms and restore a sense of touch.
Nanotechnology
The Incredible Shrinking Therapeutic
Nanotechnology is no stranger to the biomedical engineering field—in fact, it’s a common tool underlying many of the hottest areas in biomedicine. Yet when it comes to nano’s clinical potential, it hasn’t exactly reached its promise yet. That, says Mauro Ferrari, president of The Methodist Hospital Research Institute and the Houston, Texas-based Alliance for NanoHealth, may finally be changing. There are now hundreds of companies in the nanomedical space, and even pharmaceutical giants such as Pfizer and AstraZeneca have begun investing in the area. Last year, in cancer treatments alone, there were at least 117 nano-based drugs making their way through various stages of clinical testing. A sense of genuine change hangs in the air, and though it is too early to predict for sure, as Ferrari says: “Nano is coming into maturity.”
- Drug Carriers: First will be nanodelivery. It’s the farthest along, and the most likely to have immediate benefits. Multiple clinical trials are already underway, including Cambridge startup BIND Therapeutics’s drug-laden nanodelivery system, accurins, developed by the Massachusetts Institute of Technology’s (MIT’s) Bob Langer. After closing nearly US$1 billion worth of deals with companies such as Pfizer, Amgen, and AstraZeneca, BIND has begun phase 2 clinical trials with therapies to treat several cancers. Adding to the momentum is the emerging idea of systems nanotechnology, thanks in part to Sangeeta Bahtia’s work and, going forward, the NanoDoc project (see “Crowdsourcing the Nanoswarm”), which hopes to increase nanodelivery efficacy through swarming behavior and smart particle systems. “This year,” Ferrari predicts, “we are going to see breakthrough results in the clinic for some nanotechnologies that have to do with drug delivery.”
- Diagnostics: Nanodiagnostics is the next area on experts’ radars after delivery. There are two big moves happening here, according to Justin Gooding, codirector of the Australian Centre for Nanomedicine. Using particles that travel through the body as a sensing system and report back information is one area to keep an eye on, he says, but more exciting is the idea of massively parallel molecular sensors—chips, for instance, which can detect rare events in the blood that might otherwise escape detection. “Every device right now that you can think of is giving us average information,” Gooding says. “But biology occurs at the level of individual molecules, and a malfunction at that level can provide information about a disease.” Finding those events in parallel analyses could be the key to detecting rare-event diseases such as cancer. People have already been working on existing single molecular detectors, he says, and high-throughput expansions on this idea are unfolding now.
- Wired Skin: Even though it’s a little farther behind on the road to clinical impact, researchers will want to keep their eyes on nanoelectric scaffolds for engineered tissues over the coming year. The idea is simple—instead of using a general gel matrix to support cells while they develop, scientists can create a nanowire mesh with built in transistors that support the tissue on a scaffold designed to record whole tissue drug responses, pH changes, and general electrochemical status in real time. The pioneer in this area is Harvard University’s Charles Lieber, who began publishing on the concept several years ago. His laboratory has already successfully cultured heart, muscle, and neural cells on such nanoelectric scaffolds, and as he suggests in his writing, this might ultimately lead to a new experimental paradigm with which to study the interface of man and machine for regenerative medicine, neuroprosthetics, and brain mapping.
[accordion title=”Crowdsourcing the Nanoswarm”]
Large, small, loaded with cargo, crafted from gold—there are a thousand and one ways to build a nanoparticle, and every one of them changes how the particle behaves when it’s seeping out of blood vessels with a trillion of its peers. Trying to run through all of the possible scenarios to address even a single biological situation takes a tremendous amount of time, money, and manpower.
So why not tap into the crowd? That’s the approach adopted by Sabine Hauert and her principal investigator, Sangeeta Bhatia, at the MIT Laboratory for Multiscale Regenerative Technologies, in their newly released nanotech strategy game NanoDoc.
Bhatia’s laboratory specializes in designing nanoparticles for cancer treatments. Several years ago, she realized that nanomedicine had hit a wall: the field was extremely good at producing sophisticated nanoparticles, few of which were very good at getting where they needed to go to be effective. A systems nanotechnology approach was required to push the translation of these sophisticated particles into a complex collective that, when combined, acts with direction—that is, swarms.
In 2011, the laboratory published one of the first pieces on this concept, reporting on a one–two punch approach in which a first type of nanoparticle found a tumor and then signaled a second type of particle loaded with the treatment. It worked beautifully, delivering more than 40 times the dose of chemotherapeutics than its nontagged-team counterparts managed, and helped spur an emerging wave of nanosystems thinking. But still, it was just a single answer to a single biological question.
“The problem is that the possibilities are very wide, and once you solve the problem, it usually only works for one type of scenario,” says Hauert. “We really want bioengineers to be able to pose lots of different scenarios.” That’s when she got the idea to turn to the creativity of the masses.
Launched in 2013, NanoDoc walks gamers through concepts such as diffusion and smart materials, then offers up a testing ground of digital cancer cells and particle editors that let the user dream up new particle attack swarms. When such a “treatment” launches, the chosen parameters are sent back to the laboratory’s simulator, which calculates how the settings would play out according to real-life limitations.
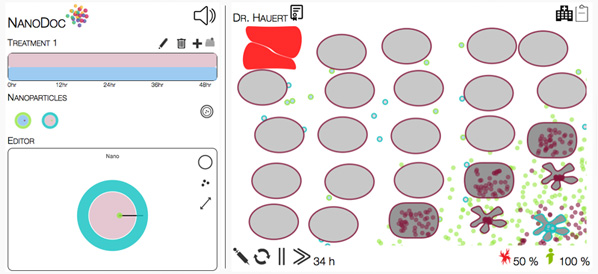
Within two weeks of its launch, 2,000 users had already worked through 40,000 scenarios. Now, as solutions pour in from the crowd, the laboratory’s bioengineers are translating the winning strategies into robotic swarms and actual nanoparticles that they will test in vitro on tumor-on-a-chip models—stops along the road, the labs hope, to smarter systems nanointerventions down the line.
[/accordion]
Informatics
Hospitals Get Smart
Information is forcing change as researchers and clinicians everywhere face an avalanche of data via better sensors, smartphones, high-throughput genomics, and global research databases. The potential for care improvement is great, but the trick is finding out how to make that information useful.
As ground zero for patient care, hospitals could be a hotbed for such research—especially with electronic medical record adoptions at an all-time high, plus a renewed emphasis on cost control in the face of health care reform. But at the same time, these institutes have been hampered by internal barriers that hinder the flow of information. From one floor to the next, one machine to the next, technologies and programs often are not compatible, and every department speaks and writes in its own medical shorthand with its own limited focus on the patient’s body. “The reality is, we are whole human beings,” says Stephen Wong, chair of the department of systems medicine and bioengineering at the Houston Methodist Research Institute. “Very few of [the caregivers] have a total picture about the disease state of the patient.”
But over the past several years, a few hospitals and health centers have broken from tradition and quietly re-engineered the way they do medicine. Most of these changes are still young, and their designers continue to refine methods and gather patient outcome data. But in many cases, the first stages are already complete, and those who masterminded them say that change is not far away.
- The Mayo Clinic in Rochester, Minnesota, has spent the last five years extracting data from electronic medical records, physiologic monitors, and incompatible test platforms, feeding it all into a large, hospital-wide data warehouse. The end result, says Brian Pickering, who worked on the project, is fully integrated data systems that follow the patient throughout their hospital stay. Essentially, what the system does, he explains, is connect all the hospital’s disparate information producers, sifting through the thousands of data points recorded on a patient in a day so that it can highlight crucial signals for doctors—or even send smartphone alerts when necessary. Pickering calls it “ambient intelligence”—unnoticeable, but ever-present. Aided by a US$16 million portion of a US$1 billion investment from the Center for Medicare & Medicaid Innovation for research into better care delivery, the clinic has just completed its installation phase. It will begin gathering patient outcome data over the course of 2014 to ensure that the changes result in improved care before it goes widely public with the concept. Meanwhile, three other hospitals, located in Massachusetts, Oklahoma, and New York, have also partnered up with the Mayo Clinic to test drive the idea in their systems.
- The Houston Methodist Hospital in Texas has created the United States’ first formal systems medicine department. Its goal is to harness bioinformatics, molecular “omics,” imaging, and systems biology data to the level of the patient. To do this, it pulls experts from their constraining specialty departments—computer science, data mining, biology, clinical, imaging, and microscopy—and combines them into an interdisciplinary team focused on patient care. “You can’t expect a physician to develop an algorithm device on new drugs or build a data warehouse,” explains Wong, who founded the department. “We’re very project-based and problem-driven.” If he has a patient with a cancer that is unresponsive to a particular treatment, he can have his team access pharmaceutical research and clinical trial datasets for known drugs (he has already partnered with various disease centers at Houston Methodist and some pharmaceutical companies to do this) and try to find a match quickly based on patient genomic profile and drug interactions. At every step, everyone contributes their differing expertise. It’s the tip of the iceberg, Wong says—an early move into the challenge of what to do with Big Data. “It’s a mindset change,” he explains. “Besides finding new uses for old drugs, we identify a couple of other areas where we can really do some quick fixes and have an impact on health care right now. This field will be growing further.”
- Surrounded by sensors, insulin pumps, continuous glucose monitors, and patients’ activity monitors and pedometers, the Joslin Diabetes Center in Boston, Massachusetts, has decided to smarten how it manages diabetes care. Teams there work to find the right predictive algorithms and analytics to distill the data into useable, automated expert support systems. According to John Brooks, president of Joslin Diabetes Center, the work is currently ongoing and will, he hopes, catalyze a movement beyond Joslin to harness the sea of numbers flooding in from wired diabetics.
Diabetes
Pancreas Proxies Hit Their Stride
Worldwide, 347 million people suffer from diabetes, and if trends continue as they are, the disease will be the seventh leading cause of global death by 2030. Many of these patients suffer from type 2 diabetes, which has spread in tandem with the obesity epidemic. But a growing number, for reasons still not fully understood, suffer from type 1 diabetes, the mysterious autoimmune disorder caused by the body’s own rejection of insulin-producing cells. It’s an easily fatal disease that demands lifelong insulin therapy and constant self-monitoring.
[accordion title=”Engineering the Microbiome”]
Glass bottles, tangle of tubes, sensors everywhere—it doesn’t look like an intestine, but as far as the roiling bacterial communities within are concerned, it’s the next best thing. For the scientists behind it, it’s the future of microbiomic engineering.
Designed by University of Guelph microbiologist Emma Allen-Vercoe, the RoboGut is a small-scale bioreactor that continuously cultures different intestinal bacterial ecologies for weeks at a time, in parallel and under precise computer-monitored parameters. In combination with high-throughput DNA sequencing technology, the whole system can maintain and analyze entire human gut communities and their natural social and metabolic behaviors.

“Microbes are, in general, a very gregarious lot,” Allen-Vercoe says. “They have a social order and a social structure.” A lot of work so far, she explains, like the Human Microbiome Project, has documented which microbes are present in human guts, but hasn’t really looked at how they behave in nature—key to the microbes’ metabolic outputs and role in health and disease.
Allen-Vercoe has already used the RoboGut to develop purified microbial treatments for C. difficile patients—literally fecal transplants without the feces. Now, she has teamed up with the Hospital for Sick Children and the University of Toronto immunologist Jayne Danska to study the gut biota of type 1 diabetic children. Theirs is part of an emerging body of work into the diabetic biome, something that experts believe will become clinically crucial in the future as evidence accumulates. A series of studies over the last few years has already reported differences in the gut populations of healthy children with genetic and immune markers of diabetes risk factors compared to kids without these markers. Last year, Danska found not only that microbial differences between male and female mice bred for diabetes correlated with their risk of developing the full-blown disease—females were more vulnerable—but that early biome transfer from male to female mice blocked diabetes development.
The next step, Danska says, is to come up with ways to manufacture reproducible core microbiota to build a normal biome and reduce inflammation in the gut in people with a high risk of developing diabetes, “to either prevent the disease altogether, or ensure that the time to disease onset becomes much, much longer—which would also be a huge breakthrough.” In short, with tech like the RoboGut to lead the microbial way, researchers might someday stop diseases such as diabetes from ever developing in the first place.
[/accordion]
Despite the development of technologies such as continuous glucose monitors and insulin pumps, managing the disorder safely and automatically—keys to a reasonably normal life for a patient—has been difficult to achieve. But big investments and innovative strategies are now beginning to pay off in the form of functional pancreatic replacements—mechanical and bio-hybrid—and daring immune-circumventing techniques, and this year, the results of the work will start coming into their own (see also “Engineering the Microbiome”).
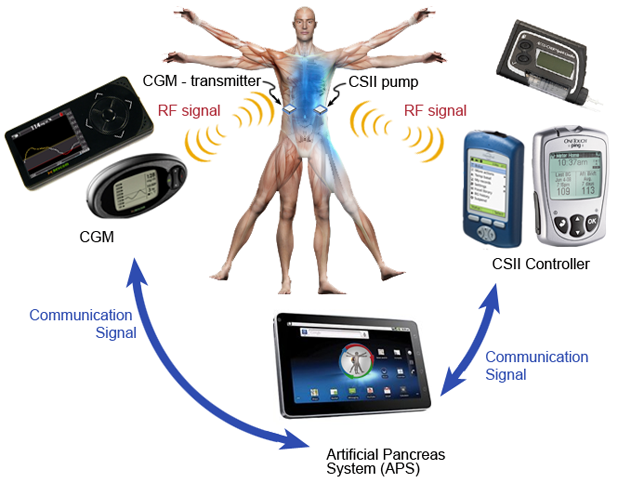
- Artificial Pancreas: Fifty years in the making, the artificial pancreas—an automated glucose-sensing, insulin-dosing smart system—is nearly here. Several inpatient studies have demonstrated that the systems are safe in controlled settings. Outpatient work has begun in the United States and Europe, and researchers are now wrestling with the finer details of their predictive algorithms as they face the unpredictabilities of patients’ daily lives. Funding continues to pour into the field from research foundations such as the JDRF (formerly the Juvenile Diabetes Research Foundation) and now from pharmaceutical heavyweights like Johnson & Johnson and Medtronic—the latter of which won U.S. Food and Drug Administration approval for its insulin suspension sensor-pump system in September 2013.
- Cell-Based Solutions: At the University of Bordeaux in France, research trio Jochen Lang, Sylvie Renaud, and Bogdan Catarg are developing what they have dubbed the Diabetachip: a bioelectric sensor made of islet cells fixed onto multielectrode arrays that can amplify and analyze cell signals in real time (Figure 2). Researchers hope their technique will sidestep the signal challenges posed by factors such as exercise and hormones, which algorithm-based systems still struggle to handle. “The cells have been shaped through half a billion years of evolution,” Lang says. “They are already integrating the signals.” The team has planned a prototype of the chip to appear this year, to serve initially, they hope, as a platform for cell screening and drug testing, then as a crucial element to automated bioelectronic pancreas stand-ins.
- Immune Evasion: Cell-based technologies such as Lang’s Diabetachip, however, will have to be used in conjunction with anti-immune technologies; otherwise, they’ll succumb to the same hostilities that produced diabetes in the first place. Traditional methods include general immune suppression, which poses a multitude of downsides. But lately, the immunological front has proven as innovative as its organ-engineering counterparts. Targeted immune suppression treatments that reduce only the anti-islet cell immune response emerged from two laboratories last year. Lawrence Steinman at Stanford University used a modified DNA vaccine in humans; Northwestern University’s Stephen Miller developed a second-generation nanoparticle treatment that reset the immune dysfunction in animal models of both diabetes and multiple sclerosis, and is planned for clinical testing this year in diabetes patients. Likewise, encapsulation techniques that protect islet cells in artificial membranes have also begun to show new promise after a recent series of small studies and a fresh infusion of investment and innovation. The California-based company ViaCyte raised more than US$10 million last year and is working toward clinical trials for a new biocompatible encapsulation technology it has developed. “That will be hugely telling,” says Julia Greenstein, vice president of cure therapies at JDRF. “Because if you could do that, and if you could protect those cells, you would demonstrate significant progress in this exciting field.”
Reproductive Health
Reproduction Gets Technical
After decades of manual technique and eyeballed assessment, assisted reproduction is going high-tech as technologies and techniques co-opted from general science give way to specialized and automated equipment designed for embryos and germ cells.
“The next decade is going to be the lab decade, and it’s been long in waiting,” says Arne Sunde, who heads the fertility clinic at St. Olav’s University Hospital, Norway. Indeed, cryogenics has already leapt forward: fast-freezing vitrification replaced slowly frozen embryos several years ago, pushing survival rates close to 100%. Now, embryo selection—the crucial key to successful in vitro fertilization—is having its own moment, as two potentially breakthrough technologies edge their way into laboratories and clinics across the world.
- Time-Lapse Imaging: Traditionally, the way to gauge an embryo’s development before implantation is to pull it from the incubator and look at it. As methods go, it’s disruptive and imperfect, but it works—about 30% of the time. In the other 70%, the embryos don’t implant or they miscarry, and that’s mostly due to unrecognized aneuploidy or another genetic abnormality. Time-lapse imaging seals embryos into an incubator with a camera that can take thousands of images of its development from moment to moment. The resulting data offer an unprecedented view of embryo development and have already shown the process to be far more complex than previously thought. International morphokinetic databases are now being compiled so that researchers can get an algorithmic handle on what constitutes healthy development. Some studies have suggested that time-lapse risk rankings can predict which embryos will succeed. “The time-lapse project is one of the hottest things going on,” says David Battaglia, a fertility specialist at Oregon Health & Science University. “There’s been an incredible flurry of interest.” But, he adds, the field needs more data from larger studies before it can truly go mainstream.
- Next-Generation Sequencing: Genetic analysis is the other way to check an embryo’s viability—but it is also expensive, time-consuming, too often requires that embryos be frozen at critical junctures, and, very simply, has never achieved its promise. Next-generation gene sequencing may change all that. The technique has already wrought transformation in fields across science, but it has only just been introduced to embryology, pioneered largely by University of Oxford researcher Dagan Wells. It can analyze multiple embryos for chromosomal, mitochondrial, and genetic diseases in as little as 16 hours and from a single cell per embryo—that means same-day testing and implantation, no freezing necessary. The first baby selected through the technique was born last summer, and some already hail the tool as a revolution. Like time-lapse imaging, however, next-generation sequencing will also need broader confirmation before it can move forward, but studies are planned for this year to provide exactly that confirmation.
Forward the Foundations
Surveying the field, so much is poised to happen this year, both in the trends we have listed here, and the many more that we could not. The entire field of biomedical engineering is afire with innovation right now, invigorated by increasingly interdisciplinary research teams, new tools, and above all—basic curiosity.
That last item on the list is fundamental. At heart, curiosity is why the brain initiatives took form, why BMI is gaining such speed, why CRISPR-Cas (see “CRISPR Transforms Genome Editing”)—which will almost certainly open up genetic engineering to even the smallest of laboratories—was discovered in the first place. In fact, the most exciting tools in bioengineering today, such as CRISPR or optogenetics, were found not because anyone was looking for them but because their discoverers simply wanted to know more about microbial immunity and ion channel function. That’s why it is so important to fund even this level of basic science and not just the headlining initiatives, says the Broad Institute’s Feng Zhang, who was involved in pioneering both tools. “And what you will see in the coming year or years,” he adds, “is that more and more of these molecular processes will get harnessed and used as molecular technologies for either treating disease or as research tools to better understand biological systems.”
[accordion title=”CRISPR Transforms Genome Editing”]
It can take at least one year and more than US$10,000 to breed a superior mouse, and that is considered fast and cheap. Or rather, that’s how much it used to cost. Now, genome editing is in the midst of one of the fastest revolutions to ever hit biomedical science, and the name of the tool that’s driving the change? CRISPR-Cas, short for Clustered Regularly Interspaced Short Palindromic Repeat and the nuclease associated with it (Cas9).
In nature, the CRISPR system is part of an RNA-guided immune system that helps microbes fight off phages by matching and attacking the viral invading DNA. But over the course of the past several years, a handful of laboratories have been quietly picking away at this system, until last winter, when four teams across the United States, Europe, and Asia almost simultaneously figured out how to transform the microbial quirk into an all-purpose genome editor light-years better than anything that has ever come before.
The tool is extraordinarily fast, easy, and cheap: with just a guide RNA and the Cas9 nuclease, the CRISPR system zeros in precisely and efficiently on the target gene and gets to work. Researchers have already tested it in everything from mouse embryos and C. elegans to zebrafish, bacteria, and crop plants—even human cells. It can quite literally add, delete, activate, and repress any gene in (so far) any genome, at will. All a researcher really needs to do is order a set of CRISPR-Cas component plasmids—US$65 at repositories like Addgene—and, in less than a week, they can have a system ready to test.
“One student can easily make a dozen new CRISPRs in one day, or automated high-throughput methods can make thousands in parallel,” says Charles Gersbach, a biomedical engineer at Duke University. “The boundaries are kind of limitless.”
“There’s no limit—and any lab can use it,” agrees Caixia Gao, a molecular agrobiologist at the Chinese Academy of Sciences. She believes CRISPR will accelerate genetic engineering across the board. Genetically modified plants, for instance, could be created through targeted tuning of amino acids here and there, rather than importing unwieldy foreign DNA from outside bacteria, making them, if nothing else, more palatable to consumers. Others foresee breakthroughs in basic gene research and even therapy—correcting genes that cause sickle cell anemia, Huntington’s, muscular dystrophy, or cystic fibrosis. Several companies, such as Caribou Biosciences—cofounded by UC Berkeley researcher Jennifer Doudna, an early pioneer on CRISPR—have already kicked off with the intent to harness the new tool for medical and biotechnological uses.
“It’s taking the field by storm,” Doudna says. “The last time I looked, there were more than 60 publications out in 2013 using this technology since January. It’s been really quite remarkable. I’ve never seen anything like this in my career.” Most of the work so far has been proof-of-concept, simply showing which genomes work with CRISPR, she says, but that phase is already winding down, and in 2014, she believes it will become a standard laboratory tool for genome editing. In other words, a new era is about to begin.
[/accordion]
“There’s an incredible value in basic research,” agrees UC Berkeley’s Jennifer Doudna, who also worked on CRISPR. “This is how scientific discovery goes, and you just cannot predict where breakthrough technology is going to come from.”