Structure-based drug design is on the front line of promising advancements in disease treatment and personalized medicine. However, the difficulties of characterizing protein structures hamper these drug development efforts. To visualize the topography of a protein, one must crystallize the protein in a solution, outside its natural environment of the human body. Finding the ideal solution in which to grow protein crystals for three-dimensional (3-D) structural analysis is an arduous process; most proteins have distinct optimal requirements for crystallization, so no standard experimental conditions are available that will work for all proteins in the human body.
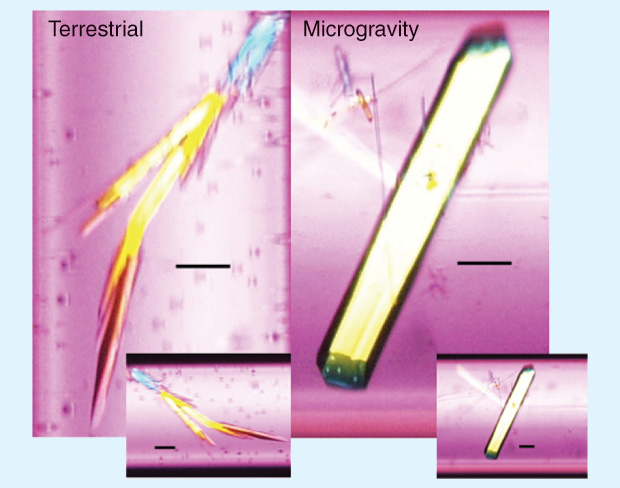
Optimizing growth conditions is paramount to growing crystals suitable for structural analysis, and, somewhat unintuitively, gravity is a potentially critical variable in this process. By performing crystal growth in the absence of gravity—that is, in space—researchers could produce more perfect crystals and, thus, acquire medically relevant data faster. One such experiment beautifully illustrates the Earth benefits of space-based research: the growth in microgravity of a protein involved in Duchenne muscular dystrophy, alone and in complex with potential inhibitors, led to the discovery of previously unknown structural features [1] in these proteins that enabled the improved design of a therapeutic inhibitor. That drug has passed through animal testing and is moving into human clinical trials.
Background: Crystallization and Analysis
The crystallization of proteins and other macromolecules allows scientists to determine structure by methods such as X-ray crystallography. Structure–function relationships are critical to understanding how molecules behave and interact in their native environment; thus, determining macromolecular structure advances basic scientific knowledge and supports commercial applications such as drug development and protein engineering.
Two major hurdles in protein and large-molecule crystallography are 1) production of adequate amounts of active, properly folded, purified macromolecules and 2) attainment of high-quality 3-D crystals. These hurdles limit our ability to determine the structures of important molecules. Although recent advances in protocols for crystal analysis [2] promise to enable crystallization of very small amounts of protein, finding each protein’s optimal conditions for growth remains difficult and time-consuming. No single method or technology is optimal for all proteins, and many structures remain elusive or produce crystals with insufficient quality for meaningful analysis. For example, membrane proteins regulate what enters and exits a cell, including drugs to treat disease and microorganisms that may cause disease. Membrane proteins constitute approximately 25% of the human proteome and more than 40% of drug targets, yet researchers have resolved fewer than 150 unique structures.

Performing crystallization in space’s microgravity environment offers one potential solution to the problem of crystal quality. Many space-based experiments have yielded larger, more well-ordered crystals, most likely because of the near absence of buoyancy-driven convection and sedimentation effects in microgravity [i.e., these gravity-dependent phenomena are functionally removed due to the free-fall nature of the orbit of the International Space Station (ISS)—the angular acceleration of its orbital path]. These improved crystals are more likely to enable structural determination (e.g., higher resolution from X-ray diffraction techniques or larger-volume crystals advantageous for neutron diffraction methods).
Preliminary EXPERIMENTS in Space
The first experiments evaluating crystallization in space (including attempts to produce higher-quality crystals) began in the 1980s on the Russian space station Mir and the U.S. Space Shuttle, well before the completion of the ISS. These experiments evaluated macromolecules including proteins, RNA, DNA, nucleoprotein complexes, and large assemblies (e.g., nucleosomes, ribosomal subunits, and viruses). The rest of this article will use the term protein (albeit incorrectly) to refer to all of these biomolecules.
The most common technique for early space-based crystal-growth experiments was vapor diffusion, in which a protein–precipitant mixture is allowed to move through a vapor space into a reservoir of more concentrated precipitant. These early investigations, when compared with terrestrial data, showed that gravity-driven convection currents affected how quickly individual molecules attached to a growing crystal, sometimes leading to irregularities and impurities. Gravity-dependent sedimentation effects also appeared to cause crystals to clump, making them unusable for X-ray diffraction. Although laboratories have produced protein crystals for more than a century, scientists still do not fully understand crystallization. Thus, these early findings not only improved crystal quality and the resulting data acquisition but also advanced the basic understanding of crystal growth by highlighting a crucial role of gravity.
Early experiments characterized well-known protein structures produced from space-based crystallization, and meticulous comparisons indicated that microgravity did not alter the conformation (i.e., the folding and final structure) of proteins [3]. These experiments used model molecules (e.g., aspartyl-tRNA synthetases, lysozyme, and thaumatin) as proof-of-principle pilot studies and to optimize crystallization conditions in space. Also, before performing structural determinations and biochemical analyses after the experiments returned to Earth, investigators used optical microscopes, time-lapse image recording, and video microscopy to analyze experiments during crystal growth. The results from these early experiments noted several beneficial aspects of crystal growth in space and subsequent analysis, including larger, well-shaped crystals with more developed features, a higher resulting diffraction limit (resolution), and a lower observed mosaicity (misalignment within the crystal lattice). On the basis of knowledge of the effects of microgravity, investigators hypothesized that the observed improvements in crystal growth and quality resulted from the slower movement of molecules during crystallization in space, resulting in a more uniform incorporation of molecules into growing crystals.
Early Successes
Early experiments aboard Space Shuttles used several facilities, and evaluations of data from the various facilities are comparable. For example, two reviews [4] analyzed data from experiments that used the Advanced Protein Crystallization Facility (APCF). The APCF experiments (which began in 1993 and gathered data from six Shuttle missions) included 474 samples from 46 proteins. Data from these experiments supported the overall conclusions from early experiments: lowered mosaicity, large crystal volume, and more defined morphology. Table 1 shows major conclusions from the APCF data.
Table 1. Finding From APCF Data [4].
| Conclusion | Occurrence in Studied Proteins (%) |
|---|---|
| Lower mosaicity/misalignment (a) |
57 |
| Larger crystals | 65 |
| Better morphologies | 67 |
| Higher diffraction limits | 52 |
| Determination of better 3-D structures | 24 (b) |
| Fewer defects, such as twinning | 50 |
(a) Equal mosaicity to ground-produced crystals was observed as well, but no crystals showed higher mosaicity than the ground counterpart.
(b) Decreased percentage compared to the 52% above is due to results from crystals that were of higher resolution yet not stable enough for data collection.
Further analysis showed that crystal quality also improved during later missions, presumably because of both optimized space-based experimental conditions and advancements in approaches and instruments used to determine structure. As in ground studies, crystallization success depends on many chemical conditions (e.g., pH, buffer types, protein concentration, ionic strength, precipitating agent, and temperature), all of which must be optimized to produce high-quality crystals. Repetition thus expectedly increased the success rate (defined as higher diffraction limits) as well: 29% for proteins assayed only once, 65% for twice, and 78% for three repetitions.
Data further showed that the concentration-depletion zone during crystallization appears to be more stable in microgravity—indicating lower local supersaturation around each crystal—although later studies suggested that this reduction might be negligible. Crystals in solution were also more regularly spaced and showed higher nucleation probability. Overall, the APCF data showed that the diffusive conditions of microgravity are likely to be the key factor, resulting in improved crystal growth (compared with other parameters such as isoelectric point or crystal solvent content).
Unfortunately, despite these informative data, many APCF experiments yielded incomplete crystal growth because of short mission duration. In fact, the first 15 years of experimentation in space produced relatively few results because of hardware limitations, the relatively short duration of Shuttle flights (usually fewer than ten days), and other factors such as infrequency of Shuttle missions, small available experiment volume, limited sample capacity, uncertainty in launch schedule, and temperature instability. The opening of the ISS research platform promised to address many of these problems—as well as providing a platform for a variety of other drug advancement studies (see “Additional Drug Development Initiatives: A Case Study on Drug Repurposing”).
[accordion title=”Additional Drug Development Initiatives: A Case Study on Drug Repurposing”]
by Timothy G. Hammond and Holly H. Birdsall
In his landmark article “Mining for Therapeutic Gold,” Dr. Francis Collins, the director of the National Institutes of Health, discussed the myriad difficulties of developing new drugs: failure rates of up to 95%, 13 years from target selection to approval, and a cost of US$1 billion. A promising alternative that complements the existing approach to drug discovery is to repurpose currently approved drugs for new diseases. One of the best examples of this is AZT, which was developed as a cancer drug but dropped for lack of efficacy. However, AZT was repurposed as the first antiviral drug effective against HIV, and that discovery changed the course of life for millions. Dr. Collins proposed a partnership between government, industry, and academia to pursue drug repurposing.
A fundamental element to successful drug repurposing is to define those conditions in which approved drugs may manifest new, unexpected activities. For example, it is well established that the environment in which a drug acts has profound effects on its efficacy. In the spirit of Dr. Collins’ goal, we seek, through microgravity studies, to make a valuable and unique contribution to drug repurposing by exploiting the novel environment provided by microgravity. To realize this vision, the repurposing effort must be cost-effective and leverage the expertise of both government and academia. We have thus partnered with NASA and the Center for the Advancement of Science in Space to use spaceflight to induce yeast to adopt metabolic profiles that effectively model many of the features of tumors only during microgravity culture conditions.
Chemogenomics is the systematic screening of chemicals against individual drug target families to identify novel drugs and/or drug targets. In a current project that uses the International Space Station research platform, we are using this approach to identify the cancerkilling pathways of Metformin, a drug currently used to treat diabetes. Metformin is a widely used, potent, first-line oral therapy for type 2 diabetes. More recently, Metformin has been found to also protect against certain types of cancer, reduce cancer-related mortality, and even have direct tumoricidal effects, particularly toward cancer stem cells. Given its known minimal toxicity and safety profile, Metformin is poised to be rapidly moved into clinical trials for anticancer therapy via the U.S. Food and Drug Administration 505(B)(2) repurposing pathway once the mechanisms of action are defined.
Studies to define the metabolic pathways for Metformin’s anticancer capabilities have yielded a wide variety of potential mechanisms without a definitive answer. Two key variables appear to complicate the analyses. The various human and murine cancer cells used as targets have differing genetic dysregulations underlying their oncogenesis. Thus, each cancer line presents different metabolic abnormalities as potential targets for Metformin. Second, new data suggest that Metformin’s effects on cancer cells may be highly affected by hypoxia. Cancer cells that are vulnerable to Metformin, when grown in well-oxygenated cell cultures, become resistant to Metformin when grown as tumor implants in mice, where the tumor core tends to be hypoxic. The hypoxic center of tumors is a difficult microenvironment to model in the laboratory in a ground-based environment. This makes it hard to use cultured cancer cells to set up appropriate models to evaluate the pathway(s) of Metformin activity.
Yeasts are ideal for the chemogenomic study of Metformin pathways for several reasons:
- Yeasts share a high degree of genetic and biochemical homology with human cells, making it likely that the pathways identified will have clinical relevance. We have already demonstrated that yeasts are susceptible to Metformin in doses that can be generated in aqueous solutions. In the assays for our current project, yeast will divide up to 20 times in the presence of Metformin, which are many more divisions than cancer cells would undergo in a typical proliferation inhibition assay. Thus, there is an order of magnitude better opportunity to evaluate the quantitatively modest but biologically critical effects of Metformin on cell replication activities.
- Yeast growth is not dependent on insulin. Since Metformin appears to have potent insulin-dependent effects on cancer cells, yeasts offer the opportunity to analyze the insulin-independent effects away from insulin-dependent effects.
- Yeasts can easily be genetically manipulated to allow the identification of cellular response pathways with high fidelity of mechanistic molecular pathways. The entire yeast genome is cloned, and deletion series can be constructed in a variety of background strains, efficiently and cost-effectively, using fully automatic robotic approaches. The simplicity of the handling requirements facilitates adaption of yeast assays to spaceflight conditions.
- Cancers are a heterogeneous mixture of cells with multiple microenvironments within the tumor including altered redox potentials. Yeasts grown in the presence of a variety of different stressors can model these microenvironments and provide a homogeneous cellular target for analysis. A cell’s metabolic state influences how it responds to a drug. Yeast chemogenomic screens have already uncovered unique profiles of sensitivity for drug stresses in three cancer-relevant contexts (hypoxia, obligate respiration, and normoxia). Our current project builds on several lines of evidence, based on yeast chemogenetic experiments flown on the last space shuttle mission, STS-135, that microgravity provides a fourth cancer-relevant context, which extends and complements current models.
In summary, microgravity allows yeast-based modeling of drug pathways in ways that complement and extend current ground-based models. Our current project investigates the anticancer pathways of Metformin using an assay uniquely available in spaceflight—yeast grown in microgravity as a model of the redox state of tumor cells.
References
- [B] A. Vergara, B. Lorber, C. Sauter, R. Giegé, and A. Zagari, “Lessons from crystals grown in the Advanced Protein Crystallisation Facility for conventional crystallisation applied to structural biology,” Biophys. Chem., vol. 118, nos. 2–3, pp. 102–112, Dec. 2005.
[/accordion]
Research on the ISS
In the early 2000s, many of the first ISS experiments yielded better structural features of crystals, leading to the improvement of known structures and the determination of some macromolecular structures for the first time (e.g., wild-type mouse lipocalin-type prostaglandin D synthase [5]). A 2008 National Aeronautics and Space Administration (NASA) review of space-based research [6] noted that more than 65% of proteins flown in one series of experiments produced diffraction-quality crystals (though not all were superior to Earth-grown crystals). ISS facilities support a variety of techniques, including liquid–liquid diffusion, vapor diffusion, and dialysis. The ISS also now includes facilities with robotic sample handling, increasing consistency in results.
As in pre-ISS experiments, many crystals showed improved morphology, larger volume, higher diffraction limit, and higher homogeneity, demonstrated by multiple experiments—including those with model proteins (a reproduction of early proof-of-principle and optimization experiments). Resulting electron density maps from crystals grown in space often contained more detail than ground analyses, allowing researchers to determine more accurate 3-D models, and less mosaicity was again consistently seen. In other experiments, some proteins showed up to 40% more intensity in the diffraction process and up to an 80-fold volume increase.
Recent publications on crystal growth on the ISS have begun to rigorously evaluate the quality of space-grown crystals compared with ground controls. One report [7] showed that out of 12 hematopoietic prostaglandin D synthase crystals (produced using varied crystallization conditions), two showed improved formation (size and shape) and eight showed improved resolution and decreased mosaicity/misalignment when grown in space (in relation to those grown on Earth under identical conditions). This study has yielded new anti-inflammatory and anti-allergy drug candidates.
In another recently published series of experiments [8] that had identical ground and space experimental conditions, groundbased studies failed to produce one of the structures produced in space. These experiments also showed that varying experimental conditions, such as crystallization method, sample homogeneity, and precipitant, affected the success of crystal growth (as defined by the production of crystals, greater resolution, and less mosaicity), indicating that any or all of these conditions might be relevant to optimizing space experiments.
The results from space-based crystallization repeatedly attain higher-quality crystal structures. After its final assembly in 2011, the powerful ISS U.S. National Laboratory research platform became available for exploitation in this field, and many projects promise to further show the utility of the ISS for such work and produce medically relevant results, accelerating the translation of structural data to the pharmaceutical market. Opportunities for protein crystallization experiments on the ISS abound, and stakeholders such as NASA, CASIS, and other international space agencies and associated organizations will pursue this research as we advance into the decade of ISS utilization.
References
- K. Aritake, Y. Kado, T. Inoue, M. Miyano, and Y. Urade, “Structural and functional characterization of HQL-79, an orally selective inhibitor of human hematopoietic prostaglandin D synthase,” J. Biol. Chem., vol. 281, no. 22, pp. 15277–15286, Mar. 2006.
- Y. Inokuma, S. Yoshioka, J. Ariyoshi, T. Arai, Y. Hitora, K. Takada, S. Matsunaga, K. Rissanen, and M. Fujita, “X-ray analysis on the nanogram to microgram scale using porous complexes,” Nature, vol. 495, no. 7442, pp. 461–466, June 2013.
- B. Lorber, “The crystallization of biological macromolecules under microgravity: A way to more accurate three-dimensional structures?” Biochim. Biophys. Acta, vol. 1599, nos. 1–2, pp. 1–8, Sept. 2002.
- A. Vergara, B. Lorber, A. Zagari, and R. Giegé, “Physical aspects of protein crystal growth investigated with the Advanced Protein Crystallization Facility in reduced-gravity environments,” Acta Crystallogr. D: Biol. Crystallogr., vol. 59, no. 1, pp. 2–15, Jan. 2003.
- K. Inaka, S. Takahashi, K. Aritake, T. Tsurumura, N. Furubayashi, B. Yan, E. Hirota, S. Sano, M. Sato, T. Kobayashi, Y. Yoshimura, H. Tanaka, and Y. Urade, “High-quality protein crystal growth of mouse lipocalin-type prostaglandin D synthase in microgravity,” Cryst. Growth Des., vol. 11, no. 6, pp. 2107–2111, June 2011.
- C. A. Evans, J. A. Robinson, J. Tate-Brown, T. Thumm, J. Crespo-Richey, D. Baumann, and J. Rhatigan. (2009). International Space Station science research accomplishments during the assembly years: An 2000-2008. [Online]. NASA Johnson Space Center, Houston.
- H. Tanaka, T. Tsurumura, K. Aritake, N. Furubayashi, S. Takahashi, M. Yamanaka, E. Hirota, S. Sano, M. Sato, T. Kobayashi, T. Tanaka, K. Inaka, and Y. Urade, “Improvement in the quality of hematopoietic prostaglandin D synthase crystals in a microgravity environment,” Synchrotron Radiat., vol. 18, no. 1, pp. 88–91, Jan. 2011.
- K. Inaka, S. Takahashi, K. Aritake, T. Tsurumura, N. Furubayashi, B. Yan, E. Hirota, S. Sano, M. Sato, T. Kobayashi, Y. Yoshimura, H. Tanaka, and Y. Urade, “High-quality protein crystal growth of mouse lipocalin-type prostaglandin D synthase in microgravity,” Cryst. Growth Des., vol. 11, no. 6, pp. 2107–2111, June 2011.



