As much as we know about the vitamins, minerals, and types of exercise important to promoting good muscle health, many fundamental questions remain about skeletal and cardiac muscle:
- What do the basic muscle components look like in real time in a living person?
- How do the signaling pathways between muscle proteins fit into the overall interacting network required for muscle movement?
- How do the muscle microstructure, dynamics, and pathways differ in a healthy person compared to someone who has a muscular disorder?
- Are there ways to identify susceptibility for some dangerous skeletal-muscle and cardiovascular diseases?
Now, innovative imaging technologies, new applications of mass spectrometry, and research using top-down proteomics are beginning to provide some important insights. Researchers hope these can propel basic science while also promoting clinical applications to help determine a person’s disease risk, diagnose disorders earlier, and possibly treat muscle-related and other diseases.
Scoping It Out

“In striated muscle, there really has not been a method for observing in live humans the basic contractile elements, which are the sarcomeres, nor for observing the dynamics of individual motor units, which are the presynaptic motor neuron and postsynaptic muscle fibers, and that is a very large motivation behind our work,” says Mark Schnitzer, Ph.D., associate professor of biology and applied physics at Stanford University.
Schnitzer, who is also a Howard Hughes Medical Institute investigator, believes that the ability to visualize these components is critical for foundational knowledge and its application to the clinic. He explains, “Sarcomere structures and lengths, muscle fibers, and motor-unit dynamics are thought to be important in many neuromuscular disorders, motor disorders, and genetic disorders of movement, but without a way of looking at them in a living person, we have no direct means of understanding what actually might be different at the elemental level between healthy humans and those with diseases or disorders of movement.”
Schnitzer’s research group collaborated with a team led by colleague Scott Delp, Ph.D., Stanford professor of bioengineering, to develop a tabletop microscope that could image individual sarcomeres in living patients [1]. “That proved the principle, but because the subjects had to come to the machine and literally stick their arms under the microscope, it wasn’t suitable for clinical investigations,” Schnitzer says. About a year ago, the teams took a big step forward when they announced that they had miniaturized the instrument into a portable unit that could come to the patient [2].
Visualization using the portable unit begins with a specially designed endoscope needle that is inserted into the patient’s muscle tissue. A handheld microscope unit attaches to the back side of the needle. Optical fibers run from a laser on a small bedside cart to the microscope unit and deliver ultrashort pulses of light to the muscle fibers. The muscle fibers respond with an intrinsic optical effect that arises naturally through a well-known process called second harmonic generation, and the backscatter of the light from that optical effect provides a real-time image of the sarcomeres.
One of Delp’s doctoral students, Gabriel Sanchez, accomplished the bulk of the massive miniaturization undertaking as part of his dissertation. “He just did a tour de force piece of work. It really shows systems engineering at its best,” Schnitzer remarks.
The miniaturization required jumping a number of hurdles. For starters, the researchers had to design the endoscope, which involved shrinking an objective to fit inside a hypodermic needle. They also had to decrease the overall size of the microscope, which demanded a range of engineering skills as well as state-of-the-art microoptics. Next, they took on the added challenge of making the device sufficiently user-friendly so that a standard technician—without training in advanced microscopy—could use it, Sanchez says. “That meant we had to solve all the problems of coupling light into fibers in such a way that you don’t have to align them. We had to provide a focusing mechanism that gives you defraction-limited performance over the entire focus range, which will allow you to search through the tissue to find the sample you want. And we had to provide an on-screen, real-time display that doesn’t need any postprocessing and maintains its optical performance.”
All the effort paid off. The microscope does everything they wanted and does everything well, Sanchez reports. “If a surgeon were to look at our images, I believe he or she wouldn’t really be able to tell our images that came out of a live subject through our needle and handheld microscope from those images that came out of a prepared and cut-out biopsy sample that’s put in a big lightproof microscope on a table. In fact, sometimes I think our microscope’s images are actually better.”
Once word started getting around about this easy-to-use, handheld microscope and its ability to clearly image muscle microstructure in living subjects, researchers from far and wide began reaching out to learn the technique or collaborate on the Stanford group’s studies of muscle function and disease. “Based on that feedback,” Sanchez recalls, “we started thinking this would be much more impactful if we provided the technology broadly so that others could do basic scientific studies and research with it.”
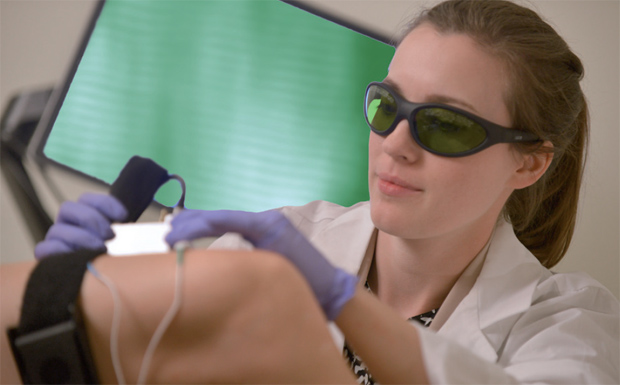
Delp, Schnitzer, and Sanchez founded Zebra Medical Technologies of Mountain View, California, which has commercialized and begun selling the newly named Zebrascope for the research market (Figure 1); Sanchez serves as the company CEO. The company is now considering developing the device for clinical use, Schnitzer says, but most of the effort today centers on building a similar microscope technology that will image cancerous tissue rather than muscle (Figure 2). “I have to be a bit vague about the details at this point, but this device is designed to detect signs of cancer using real-time imaging, and it will do this completely noninvasively,” Sanchez says. The company is planning to begin a pilot study of the new device in early 2017.
Mass Spec Eyes Muscles
Another way to look at muscles is via mass spectrometry, which has become an important emerging method for characterizing proteins. A researcher from the University of Wisconsin–Madison (UW-M) is now employing mass spectrometry to understand sarcopenia, the age-related loss of muscle mass and function common to older people. Sarcopenia typically causes muscle weakness, which can lead to falls, fractures, and often permanent disability.
“Most people see this as a natural process of aging, so sarcopenia has not been studied extensively. Nonetheless, this is a major health problem,” says Ying Ge, UW-M associate professor of chemistry and of cell and regenerative biology, as well as director of mass spectrometry in the Human Proteomics Program at the UW-M School of Medicine and Public Health. “Among people older than 80, 60% suffer from sarcopenia, and most of them experience loss of independent living. The current healthcare costs associated with sarcopenia are estimated at US$18 billion in the United States alone, so we really need to start worrying about it now.”
Ge and her research group are using a rat-aging model to uncover the underlying mechanism of sarcopenia, including the cause of the loss of muscle function, as well as to identify potential therapeutic targets to recover that lost function. In particular, they are focusing on myofilaments (also known as the contractile apparatus), mainly composed of actin, myosin, and other myofilament proteins such as troponin and tropomyosin, which are necessary for muscle contraction.
To get a good look at the myofilament proteins, Ge and her research group are using a novel systems-biology approach integrating ultrahigh-resolution mass-spectrometry-based top-down proteomics and muscle-function studies (Figure 3). Top-down proteomics comes at protein study differently than traditional “bottom- up” proteomics. In the traditional method, researchers cut proteins into small peptides, attempt to understand the role of each peptide, and then, in some cases, try to reconstruct an overall understanding of protein function. In top-down proteomics, conversely, researchers view the whole proteins [3].
“This enables us to see the important post-translational modification changes in the myofilament proteins, such as phosphorylation, which are involved in signaling and play critical roles in muscle contraction and relaxation,” explains Ge. “We need to look globally at how proteins are interacting with one another, and that’s what this type of top-down proteomics-based systems-biology approach allows us to do.”
To conduct their work, Ge and her research group developed chromatographic technologies to separate intact proteins and then use high-resolution mass spectrometry to detect and analyze these intact proteins. They also worked on methods to improve protein solubility and the sensitivity of mass spectrometry, which measures changes in phosphorylation and other molecular details.
Recently, she and her group demonstrated that myosin regulatory light chain (RLC) phosphorylation might be a critical indicator of the functional loss in skeletal muscle, which suggests that it may be important in developing therapies for sarcopenia [4]. Specifically, they discovered that high phosphorylation of RLC corresponds to higher contractility, and low phosphorylation to lower contractility. “This is critically important, because these results strongly supported a role for decreasing RLC phosphorylation in muscle dysfunction in sarcopenia and suggested that therapeutic modulation of RLC phosphorylation may represent a new avenue for the treatment of sarcopenia,” Ge says, noting that her group is preparing to publish more details of their findings in 2017.
The research group is now looking into the upstream signaling pathways that might affect phosphorylation. “If we can find perturbations to these pathways that are causing loss of muscle mass and function,” Ge continues, “then we can potentially recover the muscle contractile function in the growing number of people with sarcopenia.”
While that work continues, her group is also looking for other myofilament proteins and signaling pathways that may be associated with muscle-function loss. “We’d also like to validate our work in other animals and are now beginning studies in a rhesus monkey model, and [we] also plan to collaborate with physician colleagues on human studies.” The group looks forward to validating its work in other animals, and is now beginning studies in a rhesus monkey model and planning to collaborate with physician colleagues on human studies. Ge says, “Both the monkey and human studies will involve biopsy and blood samples, and with data-mining models we are developing with researchers in computer science, bioinformatics, and statistics, we will be tracking down the potential mechanisms involved in sarcopenia.”
She adds, “Hopefully that will lead us to discover biomarkers to determine which older adults are at risk for developing sarcopenia, and possibly to a good therapeutic target as well.”
The Heart and Beyond
Although mass-spectrometry-based, top-down proteomics has only really been available for about five years, it is also providing insight into cardiac muscle and may well extend beyond muscle to all kinds of tissues and physiological systems, says Neil Kelleher, Ph.D., Walter and Mary Elizabeth Glass Professor of Chemistry, Molecular Biosciences, and Medicine at Northwestern University in Evanston, Illinois (Figure 4, below right).

On one front, Kelleher’s research group is using the top-down approach to study proteins in mouse models of cardiac arrest as a way to identify markers of cardiovascular disease in the peripheral blood. “We use mass spectrometry and top-down proteomics because together they are the best approach for determining the atomic composition of molecules, precisely distinguishing all the forms of proteins—the proteoforms [5]—that are present, and determining how genes are processed to actually create those proteoforms,” he explains. Such proteoforms, including those with posttranslational modifications, could then potentially be used as biomarkers for disease.
As it turns out, Kelleher’s group has indeed discovered two proteoforms that correlate with heart attack risk. Both are lipid-binding proteins, or apolipoproteins (APOs), known as APO A-1 and APO C-III. To test the efficacy of the two proteoforms in predicting heart attack risk, Kelleher’s lab is currently preparing to mount a major study that will obtain blood samples from a large population of both genetically and socially diverse people and then analyze whether high levels of the APO A-1 and APO C-III proteoforms are linked to the occurrence of heart attacks. Kelleher notes, “We need to be able to state with confidence that someone’s risk is high or low based on these proteoforms, and we can’t deploy a new biomarker unless we’ve tested it on thousands of people, so we need to do this.”
While that work is under way, Kelleher is embarking on a much broader undertaking, which he calls the Cell-Based Proteome Project [6], [7]. This project will likewise use topdown proteomics, but its objective is to catalog the hundreds of millions of proteoforms present in all the different kinds of cells in the human body. To help accomplish such an ambitious goal, he and his group developed software to quickly analyze and characterize proteomes. The software, called Pro- SightPC, is now licensed to and commercialized by Thermo- Fisher Scientific.
Although work on the Cell-Based Proteome Project has just begun, Kelleher anticipates it could be completed within 10–15 years. As work proceeds, scientists will have a much better understanding of “what proteins are there and how they are modified posttranslationally,” he says, adding that this will also lead to the development of a variety of biomarkers that can portend disease risk, provide early disease diagnosis, and reveal targets for treatment, helping to make drug development more efficient as a process.
The imaging Boom
Whether these imaging technologies are used for skeletal muscle, cardiac muscle, cancer, or other health conditions, they are going to have a significant impact on basic and applied science, Sanchez predicts. “From X-ray to ultrasound to magnetic resonance imaging, imaging innovations seem to come along every decade or so. I think we’re entering the next stage of imaging technologies now.”
These new ways of viewing tissues will enable more and better diagnostic tests, improved monitoring of disease progression, and advanced treatment options, while also potentially providing healthcare cost-savings—and these changes are coming soon, Sanchez adds. “There is a big patient need, and clinicians are very open to using this type of technology, so the barriers are very low. I believe that if we do this effectively and with the right strategy, the effect of advanced imaging technologies will be felt very rapidly, even in the next five years. It will be very impactful.”
References
- M. E. Llewellyn, R. P. Barretto, S. L. Delp, and M. J. Schnitzer, “Minimally invasive high-speed imaging of sarcomere contractile dynamics in mice and humans,” Nature, vol. 454, no. 7205, pp. 784–788, Aug. 7, 2008.
- G. N. Sanchez, S. Sinha, H. Liske, X. Chen, V. Nguyen, S. Delp, and M. Schnitzer, “In vivo imaging of human sarcomere twitch dynamics in individual motor units,” Neuron, vol. 88, no. 6, pp. 1109–1120, Dec. 16, 2015.
- W. Cai, T. M. Tucholski, Z. R. Gregorich, and Y. Ge, “Top-down proteomics: technology advancements and application to heart diseases,” Expert Rev. Proteomics, vol. 13, no. 8, pp. 717–730, Aug. 2016.
- Z. R. Gregorich, W. Cai, Y. Jin, L. Wei, A. J. Chen, S. H. McKiernan, J. M. Aiken, R. L. Moss, G. M. Diffee, and Y. Ge, “Topdown targeted proteomics reveals decrease in myosin regulatory light chain phosphorylation that contributes to sarcopenic muscle dysfunction,” J. Proteome Res., vol. 15, no. 8, pp. 2706– 2716, 2016.
- L. M. Smith and N. L. Kelleher, “Proteoform: A single term describing protein complexity,” Nature Methods, vol. 10, no. 3, pp. 186–187, Mar. 2013.
- O. S. Skinner, P. C. Havugimana, N. A. Haverland, L. Fornelli, B. P. Early, J. B. Greer, R. T. Fellers, K. R. Durbin, L. H. F. Do Vale, R. D. Melani, H. S. Seckler, M. T. Nelp, M. E. Belov, S. R. Horning, A. A. Makarov, R. D. LeDuc, V. Bandarian, P. D. Compton, and N. L. Kelleher, “An informatic framework for decoding protein complexes by top-down mass spectrometry,” Nature Methods, vol. 13, no. 3, pp. 237–242, Mar. 2016.
- N. Kelleher. (2015, Apr. 15). The cell-based human proteome project. TEDx Talk. [Online].



