Traditional genetic engineering has for several decades enabled the cutting and pasting of DNA from one place to another, allowing for all kinds of developments: giving bacteria the ability to make insulin, making crops pesticide- resistant, and increasing the size of food animals, to name but a few.

Today, increasingly efficient DNA sequencing and synthesis technologies have allowed scientists to expand their prospects, and spawned the field of synthetic biology, according to Prof. Timothy Lu, associate member of the Broad Institute and director of the Synthetic Biology Group at the Massachusetts Institute of Technology (MIT). “We are no longer talking about simply cutting and pasting DNA but actually being able to program the specific nucleotide code that goes into a cell. That gives us much more control over the ‘genetic software’ that determines what the cell does.”
The still very new field of synthetic biology—a mash-up of biotechnology, molecular biology, computer science, and many other disciplines—is charging full speed ahead. Scientists are beginning to develop, simulate, test, build, and refine new ways of programming living cells for a variety of different applications. One of many examples of the growth in the field is the International Genetically Engineered Machine (iGEM) Competition, which grew from a small, month-long undergraduate course at MIT into a competition that draws 5,000 participants from universities around the world (see “Let the Games Begin! 5,000-Strong iGEM Competition Builds the Synthetic Biology Field”).
Lu compares the current era of synthetic biology to the initial decade following the 1947 invention of the transistor, an early semiconductor device. During that whirlwind decade, scientists developed the first transistor car radio and the first transistor computer. “Given how the underlying technologies of DNA synthesis are scaling up and the range of things we’re starting to be able to do, I think this era will be like the golden age of semiconductors. It’s already on a very similar and interesting track,” he says.
[accordion title=”Let the Games Begin! 5,000-Strong iGEM Competition Builds the Synthetic Biology Field”]
In 2015, the International Genetically Engineered Machine Competition (iGEM) drew 5,000 participants from 280 (primarily university) teams representing 39 countries. Those are impressive numbers, considering that iGEM began a little more than a decade ago as a month-long Massachusetts Institute of Technology (MIT) undergraduate course. That year, in 2003, about two dozen students spent their time in the short course designing biological systems to make cells blink (light up and turn off).
As time passed, that course turned into a full-fledged competition, with teams combining molecular biology and engineering concepts and techniques to create novel biological systems. Other universities soon started sending teams, word spread, more teams signed up, and in 2012, the competition had gotten so large that MIT refashioned it as an independent nonprofit organization, according to Meagan Lizarazo, iGEM vice president.

Today, iGEM’s reach continues to expand. “We have alumni numbering probably 20,000 students, and, over the last ten years, we’ve essentially moved from an educational competition for students to a program that is training the future workforce of synthetic biology,” says Lizarazo, adding that estimates indicate about 75% of current synthetic biologists have been involved in iGEM as a student, advisor, or instructor.

As examples of how the competition advances the field, she describes one team that engineered bacteria to express a gene that creates gas vesicles, thereby generating bacteria that could float; a second team that engineered bacteria able to bind to heavy metals in contaminated waste water; and a third that, inspired by these two projects, ultimately created bacteria to sequester heavy metals from water and to float so the contamination could be skimmed off the water surface. “That’s a really simple but beautiful example of building on what’s come before, and that happened because all of the participating teams have to measure, characterize, and document their projects, as well as contribute to the iGEM registry,” she says.
The iGEM registry is a linchpin of the competition. An open-access repository, the registry works much like the computer industry’s transistor–transistor logic (TTL) data books, which provide a compendium of thoroughly described integrated circuits that engineers use to create new and improved computer applications. Likewise, the iGEM registry is a collection of well-characterized biological “parts”—specifically, DNA snippets that code for discrete biological functions, Lizarazo says. Such a registry allows the iGEM teams to choose from a catalog of parts and then link them in different ways to build new systems.

Just as the TTL data books helped to transform the computer industry, Lizarazo says iGEM’s registry is helping to speed synthetic-biology developments. “The beauty of the registry is that these parts are interchangeable and modular; assembly schemes for them are available; and the teams and other users are able to put the parts together sort of like Lego bricks,” Lizarazo says.
She explains that each of the registered iGEM teams receives a start-up kit containing 1,700 of the most popular parts in the registry. Team members work on a biological system at their own university over the summer and meet up with all of the other teams at the iGEM Giant Jamboree, which has been held since 2014 at the John B. Hynes Veterans Memorial Convention Center in nearby Boston. Lizarazo remarks, “iGEM has turned into an extremely innovative education model, one that is at the cutting edge of this new field of synthetic biology, especially through the registry, where iGEM teams follow a get-and-give philosophy by getting the kit of parts at the beginning of the season and, as they create their own parts and systems, giving back into the registry at the end of the season.”
As of November 2015, the registry contained more than 15,000 physical samples of basic and composite biological parts, all with corresponding documentation and characterization. The meticulous description of the parts is one of the things that makes the registry so helpful, Lizarazo asserts. “We want to enable a user to use it without talking to the original creator of the part, so we have custom-built software that integrates the human-readable text side of it with the computer-readable database side of it, which is something that’s pretty unique and we think very important to synthetic biology as a field.”
Lizarazo describes the growth of iGEM as nothing short of amazing. “We have teams that are continuing their iGEM projects, that are commercializing their projects and turning them into start-up companies, and that are publishing their work in research journals. That is really indicative of how successful iGEM has become over the past ten years or so, and really how important synthetic biology is becoming not just to the scientific community but to the rest of the world as well.”
[/accordion]
Zeroing in on Pathogens

Research labs around the world are working on new syntheticbiology applications. During his graduate work, Lu started engineering bacteriophages for various applications, including therapeutic and diagnostic uses. Lu and fellow graduate student Michael Koeris cofounded the Cambridge-based start‑up Sample6 to commercialize the technology and applied it to test for pathogens using engineered bacteriophages to rapidly detect dangerous food contaminants.
Sample6’s first product is able to detect Listeria, the bacteria behind the 2015 Blue Bell ice cream outbreak in the United States. “What we’ve done is to engineer the phage so that when it detects Listeria, it injects a genetic circuit into that bug, which causes it to make luciferase. Because luciferase is an enzyme that generates light, we can then use a very simple luminometer device to detect the light and easily measure whether a sample is infected,” Lu explains.
Koeris, vice president at Sample6, describes the product (called DETECT) another way: “Basically, we’re using the phages to turn the bacteria into minifireflies so they emit light that’s detectable, even when they are in very low numbers,” he says.
The phage-based test has several advantages over current methods. Two of the most important are its speed and ease of use, Koeris says. “Traditionally, food producers have had to go through very laborious microbiological enrichment, followed by molecular methods such as a polymerase chain reaction to amplify DNA, or they have had to run an assay that detects a certain (bacteria-associated) protein, and these procedures require 24–48 hours to get to the point where the bacteria are detectable.” Because not all producers can afford to wait until test results come back before shipping their food, especially if it is perishable, a finding of contamination can lead to potential consumption of tainted food, as well as costly recalls. The Sample6 DETECT assay cuts down testing time to just six hours, so shipping can wait for the results. “That’s really meaningful for food producers and for customers because no other test has ever been able to do that.”
In addition, the company’s assay can be completed successfully and conducted robustly on site by individuals with minimal training, Koeris explains, noting that other tests often require molecular biologists or chemists to run them. Sample6 got its first paying customers in the fall of 2014 and already has 17 food producers using its assay at 40 sites. The company’s next step is to develop similar tests for Salmonella and Escherichia coli, two other common food pathogens.
While that work is ongoing, Lu is conducting additional research to rapidly detect the presence of specific pathogens not in food, but rather in patients themselves. If successful, such a test would reduce the need for the broad-spectrum antibiotics doctors sometimes prescribe when they aren’t sure what microbe is causing an illness. “We’ve already demonstrated in our lab that these engineered phages could actually work in human samples, such as blood,” Lu says, “and we’re really interested in trying to develop a pipeline of assays that could provide a very rapid readout within several hours and that would be applicable in clinical settings.”
He adds that the clinical project is currently at the proof-ofconcept stage, and, if all goes well in obtaining the necessary U.S. Food and Drug Administration approvals, a product could be available commercially in several years.
Cues from the Gut

Many labs, including Lu’s, are looking into possibilities surrounding the microbiome, the enormous collection of bacteria living in and on the body of every human being. Lu explains, “We believe that the microbiome represents a really interesting way of 1) studying human disease and 2) trying to treat human disease.”
Jeffrey Way and his research group at the Wyss Institute for Biologically Inspired Engineering at Harvard Medical School are interested in using synthetic biology and the microbiome to detect disease. Specifically, they are working on a bacterial sensing system designed to pick up different disease states (pictured below). The project includes several players. One is the lacZ gene in E. coli, a normal bacterium in the large intestine. Researchers often use the lacZ gene as a so-called reporter because when it is activated, it reports its “on” status by very obviously changing the color of the substrate in a standard agarplate lab test.
The next two players are proteins, known as cI and Cro, from a bacteriophage. The researchers place the proteins next to the lacZ gene. When the cI protein is turned on, which is the normal state, the Cro protein is turned off, and the lacZ gene is shut down. Conversely, when the Cro protein is turned on, the cI protein is off, and the lacZ gene becomes active. The final participant in the project is a trigger, which activates the Cro protein and, in doing so, flips on the lacZ gene.
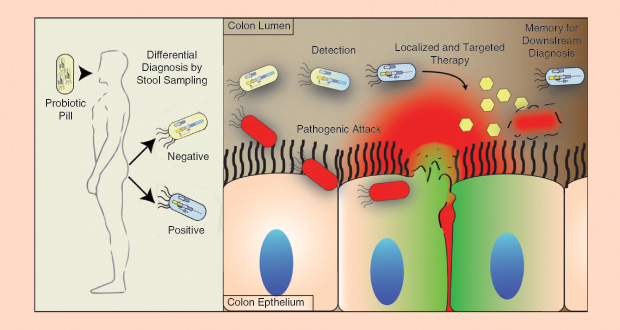
Using these four players in a mouse model, Way’s group has shown that environmental exposure to a trigger—in this case, anhydrotetracycline in the mouse’s drinking water—can be detected. “If the bacteria experience something when they’re in the gut—even if it’s temporary—when they get pooped out, you can plate them on an agar plate to see if the cI/Cro states have changed,” explains Way. This suggests that such genetically engineered systems have the potential not only to detect exposure to this particular trigger but also to identify exposure to other triggers.
One possible application is travelers’ diarrhea. Many different bacteria can cause this debilitating illness, but, if doctors had access to a genetically engineered system like Way’s, they might be able to pinpoint the cause and prescribe a specific treatment to fight it. One of Way’s major research supporters is the U.S. Defense Advanced Research Projects Agency (DARPA). DARPA is interested in travelers’ diarrhea because it is common among soldiers who are stationed overseas and also because “it has big applications across the board and major impacts on the economy,” Way explains.
Beyond diseases, such a biologically engineered bacterium could also be designed to distinguish different chemicals. “You can imagine measuring exposure to toxic chemicals on the skin,” Way elaborates. “You can imagine soldiers spraying explosive-sensing bacteria over a field to pick up land mines or unexploded bombs, and there are bacteria that can pick up nitric oxide, which is a breakdown product of TNT.”
Lu has multiple ideas, too. One is to genetically modify normal gut bacteria to contain a variety of ultrasensitive and highly specific sensors associated with various disease states, including bleeding and cancer biomarkers. “We will then be hooking up the sensors into some sort of internal circuitry inside of the bacteria so that the circuits can read what the sensors are doing, integrate multiple inputs, and make a decision about whether those inputs actually represent some disease state that you care about.”
Lu’s group has already solved many of the technical challenges inherent in building efficient circuits. The next major hurdle for this project is that of getting approval, and it may be the largest stumbling block of all. Lu observes, “This is a whole new class of therapeutics, and it has a whole new set of questions: How do you do clinical studies on these? How do you dose these? How do you manufacture these?” He helped cofound a second Cambridge- based company, Synlogic, to take on that task. It is now focused on developing synthetic biotic therapies for the treatment of inborn errors of metabolism, the rare metabolic conditions of urea cycle disorder and phenylketonuria, and possibly for more common disorders, such as inflammatory bowel disease.
Combining Synthetic Biology and Nature

Researchers at Radiant Genomics based in Emeryville, California, are also thinking outside the box. This group is using synthetic biology as a new approach to natural-product discovery, according to molecular biologist and company cofounder Jeffrey Kim.
Historically, researchers have discovered and isolated compounds, such as new antibiotics and other biologically important small molecules, from natural sources via microbial cultivation and chemical extraction. As time went by, however, research groups were turning up fewer new compounds. The problem wasn’t that there weren’t any more natural products; the problem was the strategy, Kim says. Traditional cultivation-based studies were focusing on only about 1–5% of total biodiversity because those microbes were the ones that were relatively easy to grow in a laboratory setting (pictured below). Consequently, they were looking in the same place over and over again and, not surprisingly, finding the same things.
Kim and his group are using a synthetic biology methodology that gets beyond that 1–5%. Using insights from studies showing that such natural products are encoded by relatively conserved genetic systems, they have employed genomics and sequencing to identify the enzymes involved in those systems and replaced the cultivation— as well as much of the wet lab work—with in silico sequencing and data analysis. “This gives us the capacity to do discovery on a much larger scale and for very little cost and effort, because we are taking advantage of advances in next-generation sequencing, scalable computing, and synthetic biology,” he explains.
In developing the new approach to natural-product discovery, Kim’s group set its sights on the soil, which he describes as an untapped treasure trove of natural products. “Soils are incredibly valuable environments to look for natural products and chemical diversity in microbes, but, up until recently, it was very challenging to get high-quality genomic data from soils because they are so complex,” he says. Besides using low-cost sequencing, his team built molecular-biology and data-analysis tools as well as bioinformatics pipelines that were able to examine long clusters (100 kilobases or more) of molecule-encoding genes in ways that simply weren’t possible previously. “We spent a lot of time developing downstream infrastructure— and this is where synthetic biology comes to play—that allows us to engineer and consistently heterologously produce natural products encoded by these groups of enzymes.”
In operation since 2012, Radiant Genomics currently does natural-product discovery for its clients, who approach the company with requests, such as naturally encoded analogs of target compounds. The company then finds gene clusters that encode the desired molecules and produces selected compounds at titers sufficient for immediate downstream testing. “The client takes it through the next steps, which include field testing or clinical trials,” Kim says.
One of the company’s main supporters is the U.S. National Institutes of Health. “They’re excited about the prospect of a new drug-discovery platform that is hyper-efficient,” Kim says. “This allows natural products to basically be re-explored, using more efficient infrastructure that provides access to untapped pools of genetic diversity. There is still a great deal that needs to be discovered.”
Many Possibilities
Although the field of synthetic biology is still in its infancy, it is proceeding at breakneck speed. Part of the reason for the explosion in research is that technologies have advanced to a point at which even small groups of scientists can make important contributions and become a force of innovation. Kim notes, “Through synthetic biology, we are going to see a lot of small companies developing products that impact a great deal of people.”
Another trend is the attempt over the past couple of years to migrate the same sort of tools and technologies from microbes, where synthetic biology projects currently focus, and into mammalian cells, including human cells, according to Timothy Lu. “What that is going to enable, I believe, is a collection of new sorts of gene and cell therapies for complex human diseases, such as neurodegeneration or cancer.”
As an example, he points to gene therapies that use viruses to deliver genes that are either missing or not fully functioning. “None of the existing gene therapies can be turned on or off, and none of them really has a self-regulating, self-adaptive nature associated with it, so one of the things we’re trying to do is to develop genetic circuits for gene therapy or cell therapy that can sense their environment and the disease state, and respond by making the correct set of outputs, which might include delivering a drug.” Although it’s still early in the process, Lu believes that working systems of this type will become available from research groups within the next five years or so. “This is an exciting frontier of research that’s going to happen.”
The possible applications of synthetic biology are nearly endless, adds Way. “Once you start thinking about potential applications, ideas just pop into your head, and you realize that the only thing that’s limiting is your creativity.”



