Innovations in microscopy provide new information on biological structures and activities, opening avenues for novel disease therapies
Microscopes have come a very long way since the 1600s when Henry Power, Robert Hooke, and Anton van Leeuwenhoek began publishing the first views of plant cells and bacteria. The major inventions of contrast, electron, and scanning tunneling microscopes didn’t arrive until the 20th century, and the men behind them all earned Nobel Prizes in physics for their efforts. Today, innovations in microscopy are coming at a fast and furious rate with new technologies providing first-time views and information about biological structures and activity, and opening up new avenues for disease therapies.
Some of those recent advances include building on the development of expansion microscopy to provide previously unseen detail of biological molecules; improving two-photon imaging to yield images of neuronal firing in live animals’ brains as they engage in an activity; and designing the optic hardware and imaging software in concert to get high-quality images for a wide range of applications.
Expanding the view
One of the major new discoveries in the past decade is that of expansion microscopy.
“For the first 300 years or so of light microscopy, the way you saw something small was you took an image and made the image bigger, but in early 2015 [1], we announced to the world that you could take an object and make the object bigger,” said Ed Boyden, Ph.D., who led the research group behind expansion microscopy (Figure 1). Now a professor in neurotechnology at MIT’s McGovern Institute and a Howard Hughes Medical Institute investigator, Boyden and his lab have progressed the technology to expand biological samples and expose the presence of biologically important proteins in places where they have never been seen before.
The whole idea stemmed from a limitation in light microscopy: Users have to add stains or fluorescent labels to visualize transparent objects, but it is not always successful for key molecules in biological specimens, Boyden said. “Biomolecules are nanoscale objects that communicate over nanoscale distances by touching each other and altering each other chemically, and what that means is they are often too jam-packed together for the labels to bind. So, one original motivation for expansion microscopy was to see if we could pull the biomolecules apart in an even way in order to make more room for the labels to bind,” he recounted.

Figure 1. Ed Boyden, professor in neurotechnology at MIT’s McGovern Institute and a Howard Hughes Medical Institute investigator. (Photo courtesy of Justin Knight.)
Their 2015 paper [1] demonstrated that the basic principle of expansion microscopy was possible using custom-made reagents, and within a year, the researchers were able to swap out those reagents for off-the-shelf chemicals. “And then it was off to the races with at least three lines of inquiry: extending the chemistry of expansion microscopy to visualize more kinds of biomolecules, such as nucleic acids; improving the resolution by making expansion factors that are bigger; and expanding different kinds of specimens, including human clinical specimens,” Boyden said. The MIT group and others have had success with all three.
He and his team have now progressed the technology further with so-called expansion revealing [2], a method that gives new insight into proteins (Figure 2). Currently, the typical method for labeling proteins uses fluorescent-dyed antibodies specifically chosen for binding proteins of interest, Boyden recounted. The problem is that antibodies are rather large, and cannot reach their binding sites on tightly packed proteins. “Consequently, those proteins are effectively invisible. But if we could pull the proteins apart, labels would have room to bind, and the proteins would then become visible,” he said. To accomplish that, his group developed a four-step process that: 1) applied handle-like anchors to all the proteins; 2) created and added, throughout the specimen, a mesh of polymer, specifically polyacrylate, which is highly charged; 3) softened the specimen with heat and detergent; and 4) added water, which caused the charged polyacrylate mesh to swell and neatly expand the proteins away from each other without damaging them. The stains or fluorescent antibodies could then permeate and bind.
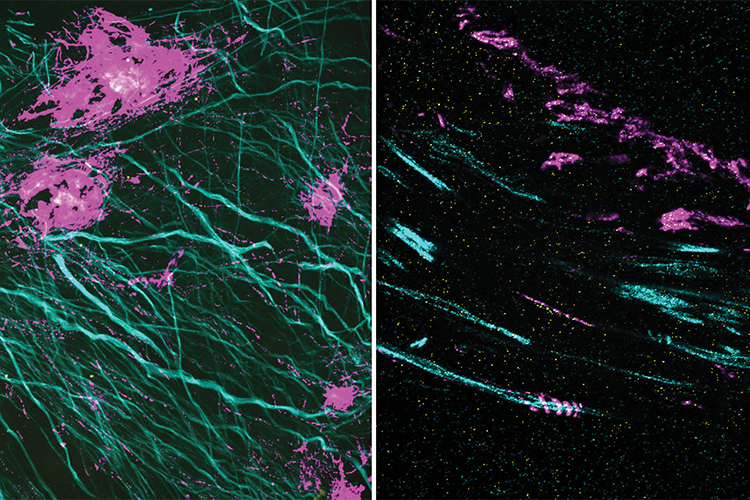
Figure 2. Expansion microscopy literally expands samples, which allows room for labels to bind, and provides detailed views of often never-before-seen structures. A new extension of that work, called expansion revealing, provides new insight into proteins, such as the amyloid-beta peptide that is linked to the pathology of Alzheimer’s disease. In these two images, the magenta signal is the amyloid-beta nanostructure, revealed by post-expansion staining. The left image shows the thread-like patterns of amyloid-beta nanoclusters, and the right image shows the helical structure of amyloid-beta, something that could not be seen with previous techniques. (Image courtesy of Zhuyu Peng and Jinyoung Kang.)
Expansion revealing worked so well that they were able to detect proteins, and “at places where they might be very important, but previously were invisible because they could not be labeled,” Boyden said. That included beta amyloid peptide, which has been linked to Alzheimer’s disease, as well as additional proteins within the brain.
His group is now refining expansion microscopy to “potentially get it down to nearly atomic scale,” while also building a multiplexing toolbox that will allow labeling of a large range of biological molecules. “My hope for expansion, looking 5 or 10 years out, is that it could help produce a map of molecules that is detailed enough to help us understand life itself,” Boyden said.
In the meantime, he is thrilled to see so many other research groups using expansion microscopy for their own work. “Currently I would estimate that more than a thousand research groups are now applying expansion microscopy, and the last I checked, they have produced more than 450 studies on everything from the human kidney to microtubules, the nucleus, chromatin, motor proteins, and viruses,” he said. “Expansion microscopy is spreading very rapidly, and I am excited that it could be applied to any biological or medical context.”
Neuronal activity—live
Two-photon fluorescence microscopy had its start back in 1930 when future Nobel Prize-winning physicist Maria Goeppert-Mayer showed mathematically that a molecule could absorb two low-energy photons simultaneously and reach an excited energy state. Since then, the development of powerful lasers as well as a variety of sensitive and specific fluorescent markers have led to two-photon microscopes that can image 3-D submicron structures deep within optically scattering tissue and do so in intact biological specimens [3]. This development was important for many fields, including neuroscience studies that benefit from views in murky, lipid-rich, and highly light-scattering brain tissue.
This technology revolutionized 3-D in vivo imaging, and now it can do even more, thanks to a new microscope developed at the University of California Santa Barbara (UCSB). Called the dual independent enhanced scan engines for large field-of-view two-photon, or Diesel2p, it uses custom, large-scale optics to maintain high resolution (enough to see individual neurons) over a large field-of-view: approximately 25 square millimeters, which can encompass multiple brain regions [4]. Group leader Spencer LaVere Smith, Ph.D., (Figure 3) associate professor in the UCSB Department of Electrical and Computer Engineering, is a principal investigator of the UCSB-headquartered hub of the Nemonic Project [5], a National Science Foundation-funded research program (part of the U.S. BRAIN Initiative Alliance) devoted to state-of-the-art technology for multiphoton imaging.
Figure 3. Spencer LaVere Smith, associate professor in the UCSB Department of Electrical and Computer Engineering, and a principal investigator of the UCSB-headquartered hub of the Nemonic Project, which is part of the U.S. BRAIN Initiative Alliance. (Photo courtesy of Jeff Liang.)
“Calcium concentrations inside a neuron increase by orders of magnitude when they fire, and we have fluorescent dyes that glow more brightly when they bind calcium. But in order to measure the activity of neurons in intact tissue and in living animals as they are doing some sort of interesting behavior, we needed to resolve micron-level features at a millimeter-length scale, and we also needed to capture neuronal activity at sub-second time resolution,” Smith explained. “That particular combination is not well-suited for conventional microscope optics. So, we went back to the drawing board to design new optics from scratch.”
To get high resolution and speed, with a large field-of-view, Smith and his group designed tube lens systems, relays, and objective lenses that Smith described as “so large they look like they belong on telescopes,” as well as new scan-engine arms that enable independent, parameter-adjustable, and dual-region scanning from a single split laser beam (Figure 4). The resulting field of view is 5 mm × 5 mm, enough to simultaneously image the neuronal activity in multiple cortical areas, some of which are 3–4 mm apart. “And then in order to improve the time resolution,” he noted, “we brought in multiple beams and used temporal multiplexing, so we can jump across two or more different sites at megahertz rates.”

Figure 4. Smith’s group designed an instrument called a dual independent enhanced scan engines for large field-of-view two-photon, or Diesel2p, which uses custom, large-scale optics to rapidly and simultaneously image neural activity over a large field-of-view (approximately 25 square millimeters). (Photo courtesy of Spencer LaVere Smith.)
Smith and his group are using the Diesel2p to study mice that are navigating through an unfamiliar space. “There is so much more we can uncover about the brain,” he asserted. “Think about it: I can drop a mouse in a room it has never seen before and in a fraction of a second, it will escape to cover, and it does so reliably based on visual navigation and using a brain that runs on less than one watt of power. By measuring neuronal activity while mice are moving through a virtual-reality environment and manipulating neural activity by causing neurons to fire—or not—using light, we can study the patterns of neuronal activity and connectivity.” If they can understand the computational principles behind that activity, he added, that could lead to improved machine learning technology and applications, such as autonomous vehicles.
Most recently, Smith’s group also developed a microscope objective lens that fits onto standard, commercially available, two-photon microscopes, and provides a considerable upgrade. “Most objectives that people use for two-photon microscopy have working distances (the distance between the objective’s front lens and the specimen) that are maybe 1–3 mm and require water immersion,” he said, explaining that both can present problems when viewing the brain in action. His group collaborated with neuroscientist Kristina Nielsen, Ph.D., at Johns Hopkins University to devise the new objective, which is called the Cousa objective, that performs in vivo, multiphoton, high-resolution imaging, and includes a correction collar to adjust for different wavelengths and optical preparations [6].
“It’s been a fun project,” he said. “Actually, we thought it would be a one-off thing, but there’s been broad interest, and there are already dozens of labs with it.”
Stronger together
At the University of California, Berkeley, researchers are also interested in observing neuronal activity, but they are taking a different approach.
“We are using computational imaging, or the idea of designing your imaging system to get synergistic improvements by using algorithms together with optics to do things that neither one could do alone,” described Laura Waller, Ph.D., professor and Charles A. Desoer, chair of the UC Berkeley Department of Electrical Engineering and Computer Sciences. “It’s not about taking existing imaging systems and doing image processing to get better images. It’s about physically changing something in your imaging system or changing the design architecture in order to get more information through, so that you can then use an inverse algorithm or some sort of image construction technique to get back the information you’re looking for,” she said. “So, if you take the measurements in an appropriate way, you can demix the information to get your final result, even if the picture you took looks nothing like your final result.”
She and her group originally demonstrated the approach [7] by replacing a camera’s lens with a simple diffuser—in their case, the diffusers included off-the-shelf pieces of scattering glass, plastic, and even standard adhesive tape—and then calibrating and demixing the acquired information to wind up with a clear and defined image (Figure 5).

Figure 5. The research group of Laura Waller (shown in bottom image), professor and Charles A. Desoer Chair of the UC Berkeley Department of Electrical Engineering and Computer Sciences, draws from algorithms and optics advances – known as computational imaging – “to do things that neither one could do alone.” They demonstrated the approach by using a “diffusercam,” a camera that has had its lens replaced with simple diffuser, to take an image of Waller (top), and then calibrating and demixing the acquired information to get the result (bottom). (Images courtesy of Laura Waller.)
Since then, the researchers have been pursuing multiplexing with this “diffusercam” approach. Waller explained, “If you’re imaging something through the diffusercam, then your point-spread function, or how your system responds to a point in a scene, is a weird scattered pattern. That can sometimes be a bad thing, because it can hurt your signal-to-noise ratio, but it can also be a good thing, because the spreading of the information means that you don’t have to take as many measurements to get information from lots of different points.” She and her group are exploiting this idea of compressed sensing to get 3-D reconstructions from 2-D measurements.
They are also optimizing the reconstruction results by fabricating various surface shapes for the scattering elements, and determining the best diffuser location on the microscope depending on the imaging goals. They have made some improvements, she remarked, but they still have plenty more to explore.
The work has many potential applications for biological microscopy, including tracking the high-speed firing of brain neurons in live animals. “While there are 3-D single-shot methods that exist, you only have so many pixels in your measurement, so you have to choose between good resolution and big volume,” Waller said. “But with this compressed-sensing idea, you can reconstruct more pixels than you measure, which means you can reconstruct a very large 3-D scene from a normal-sized measurement, as long as the sample is sparse in some way” [8].
She and her group have already shown its capability on a miniscope—the tiny, lightweight, and open-source microscope that neuroscience researchers often use to image the brains of moving mice—and have incorporated their approach with larger microscopes to image traditional samples, including the 3-D locomotion of various fast-moving little organisms, such as water bears and the common model organism Caenorhabditis elegans, Waller said. “I think there are a lot of really interesting uses there.”
Beyond biomedical applications, she envisions pairing computational imaging with electron microscopy as a way to image the 3-D arrangement of individual atoms, something that has not been possible due to the highly scattering nature of atoms. “What we are doing is to include all the algorithms for multiple scattering in order to be able to solve these problems for relatively thick, non-crystalline samples that are fairly big and have a lot of atoms,” Waller said. “We’re not down to atomic-level reconstruction in our experiments yet, but that’s our end goal, and you can imagine that it would be super-important in physics, chemistry, and materials science to understand how a material is structured in terms of which atoms are there and where they are.”
Putting it out there
Boyden, Smith, and Waller all feel strongly about making their work available to other research groups so they can further their own areas of study. “If we build a tool and nobody uses it, what’s the point?” remarked Boyden. “Our group’s first big focus was on optogenetics [8], and those tools are now being used by more than a thousand groups and have led to clinical trials of therapies ranging from blindness to Alzheimer’s. Now, with expansion microscopy, again we have more than a thousand groups practicing it, with applications ranging across the span of biology and medicine.”
Smith is also excited to see what others can do with what he started. “There are no material-transfer agreements, no licensing—everything is open-sourced so people can use it,” he said. “Not 100% of the many thousands of neuroscience labs in the world need to do the kinds of experiments that are enabled by our technology, but there are at least dozens that do, so this was about figuring out how to make an affordable, usable instrument to enable a set of experiments that they can’t do any other way.”
Waller agreed that the overall scientific picture is a driving focus in her lab, which not only freely posts its computer code on Github, but emphasizes accessibility by designing systems with easily obtained and inexpensive hardware so other research groups can easily reproduce the imaging approach and put it to use.
In other words, Waller added, “We always say that the real success is not your shiny journal paper; it’s the 10 other people using your method to write their own shiny journal papers.”
References
- F. Chen, P. W. Tillberg, and E. S. Boyden, “Expansion microscopy,” Science, vol. 347, no. 6221, pp. 543–548, Jan. 2015.
- D. Sarkar et al., “Revealing nanostructures in brain tissue via protein decrowding by iterative expansion microscopy,” Nature Biomed. Eng., vol. 6, no. 9, pp. 1057–1073, Aug. 2022.
- C.-H. Yu et al., “Diesel2p mesoscope with dual independent scan engines for flexible capture of dynamics in distributed neural circuitry,” Nature Commun., vol. 12, no. 1, Nov. 2021, Art. no. 6639, doi: 10.1038/s41467-021-26736-4.
- W. Denk, J. H. Strickler, and W. W. Webb, “Two-photon laser scanning fluorescence microscopy,” Science, vol. 248, no. 4951, pp. 73–76, Apr. 1990.
- NeuroNex. Nemonic: Next Generation Multiphoton Neuroimaging Consortium. Accessed: Dec. 13, 2022. [Online]. Available: https://neuronex.org/projects/6
- C.-H. Yu et al., “The Cousa objective: A long working distance air objective for multiphoton imaging in vivo,” bioRxiv, Nov. 2022, doi: 10.1101/2022.11.06.515343.
- N. Antipa et al., “DiffuserCam: Lensless single-exposure 3D imaging,” Optica, vol. 5, no. 1, pp. 1–9, Jan. 2018.
- F. L. Liu et al., “Fourier DiffuserScope: Single-shot 3D Fourier light field microscopy with a diffuser,” Opt. Exp., vol. 28, no. 20, pp. 28969–28986, Sep. 2020.
- E. S. Boyden et al., “Millisecond-timescale, genetically targeted optical control of neural activity,” Nature Neurosci., vol. 8, no. 9, pp. 1263–1268, Sep. 2005.