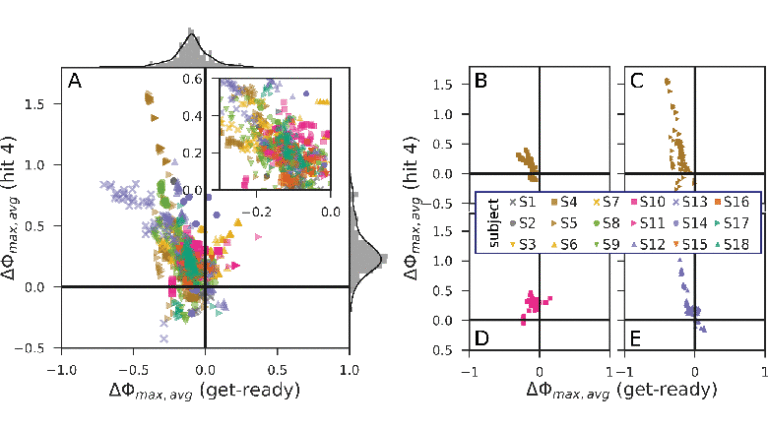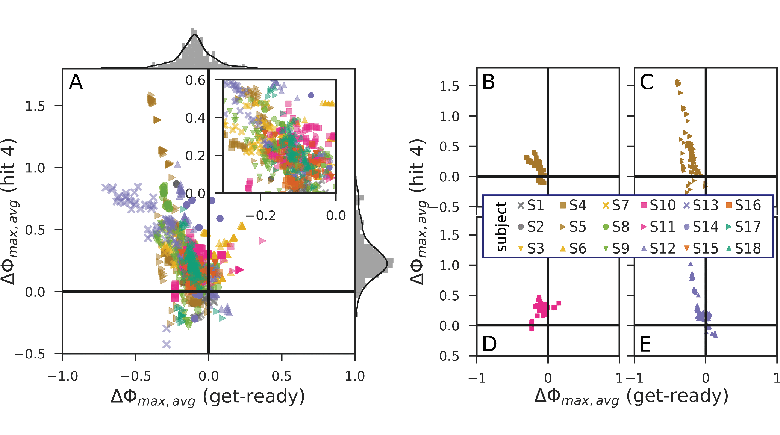
Data-driven spatial filtering algorithms optimize scores, such as the contrast between two conditions to extract oscillatory brain signal components. Most machine learning approaches for the filter estimation, however, disregard within-trial temporal dynamics and are extremely sensitive to changes in training data and involved hyperparameters. This leads to highly variable solutions and impedes the selection of a suitable candidate for, e.g., neurotechnological applications. Fostering component introspection, we propose to embrace this variability by condensing the functional signatures of a large set of oscillatory components into homogeneous clusters, each representing specific within-trial envelope dynamics. The proposed method is exemplified by and evaluated on a complex hand force task with a rich within-trial structure. Based on electroencephalography data of 18 healthy subjects, we found that the components’ distinct temporal envelope dynamics are highly subject-specific. On average, we obtained seven clusters per subject, which were strictly confined regarding their underlying frequency bands. As the analysis method is not limited to a specific spatial filtering algorithm, it could be utilized for a wide range of neurotechnological applications, e.g., to select and monitor functionally relevant features for brain–computer interface protocols in stroke rehabilitation.