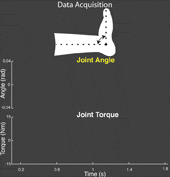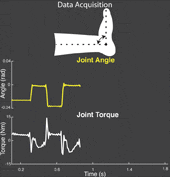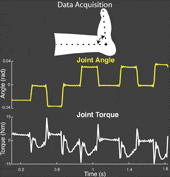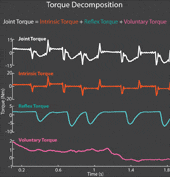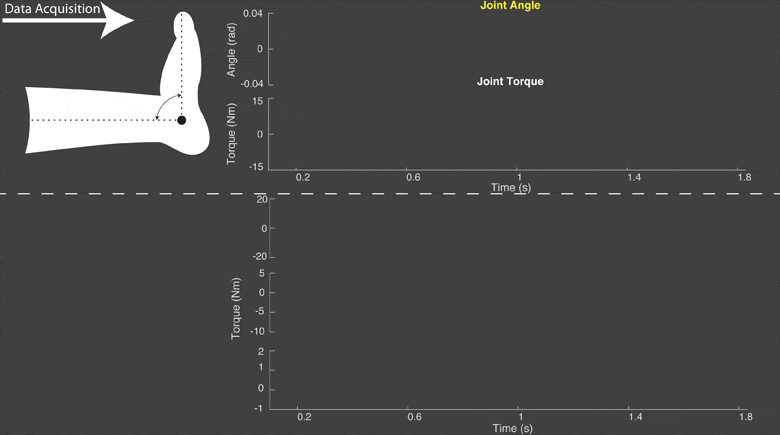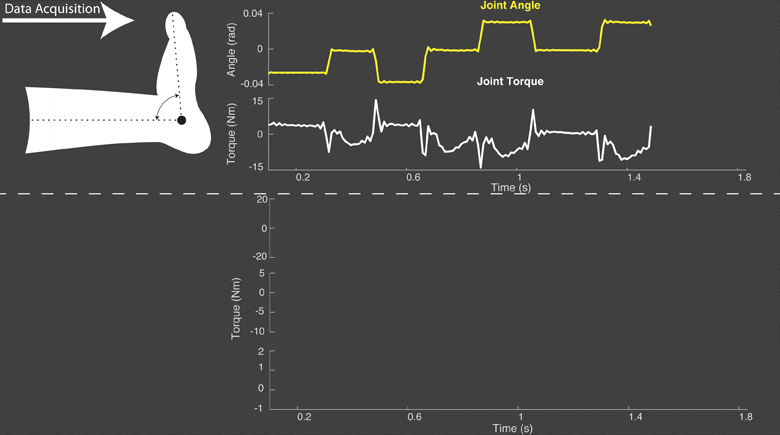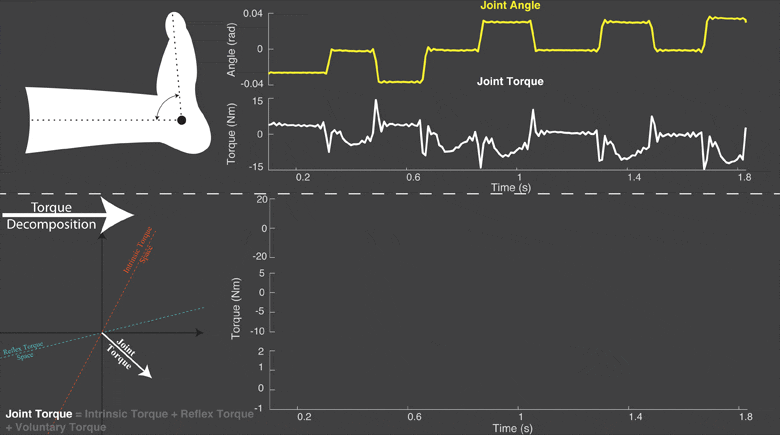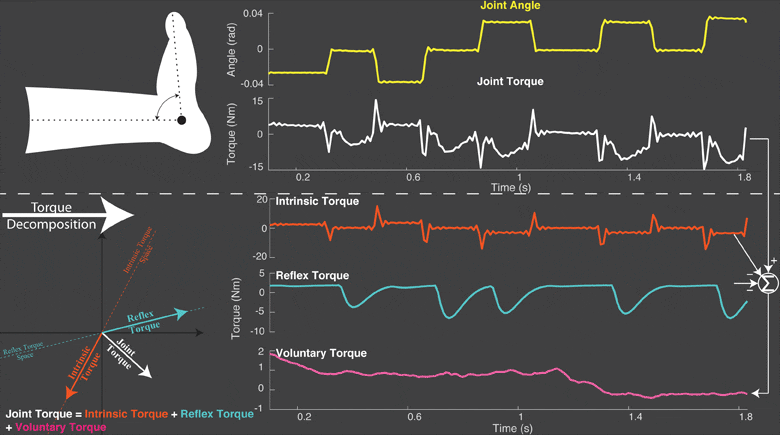
The torque developed at a joint in response to a position change arises from both neural and mechanical mechanisms. Torques initiated by mechanical mechanisms, i.e. intrinsic torque, arise from the mechanical properties of the joint and tissues. Torques generated by neural mechanisms arise from (a) changes in muscle activation due to reflex arcs, i.e. reflex torque, and (b) voluntary muscle contractions. The intrinsic, reflex and voluntary torques change together and cannot be measured individually; only their aggregate can be measured as the total net joint torque. This makes their decomposition challenging. Accurate decomposition of the torque has important clinical implications for the diagnosis and objective quantification, assessment and monitoring of neuromuscular diseases that change the muscle tone such as in spinal cord injury, cerebral palsy, multiple sclerosis, stroke and Parkinson’s disease.
The purpose of this paper is to present a Structural Decomposition SubSpace (SDSS) method for segregating the intrinsic, reflex and voluntary torques using measurements of total joint torque and joint angular position. SDSS decomposition is non-iterative and accurate. The method requires little a priori information and can deal with arbitrary coloured noise.
In the paper, we validated the SDSS algorithm using extensive simulation studies and demonstrated that it was robust to large noise conditions; it was more accurate than previously developed methods, giving estimates with lower bias and random errors. The method also worked well in practice and yielded high quality estimates of intrinsic and reflex torques from experimental data gathered from healthy human subjects.
MATLAB code for the SDSS algorithm is available from our Github repository. Run sdssDemo.m in the stiffnessID folder for a demo.