Ali Demir and H. Ertan Cetingul, Bogazici University, Turkey & Siemens Corporate Technology, USA, Volume: 62, Issue: 6, Page(s): 1478 – 1489
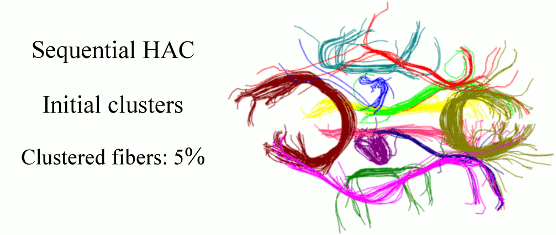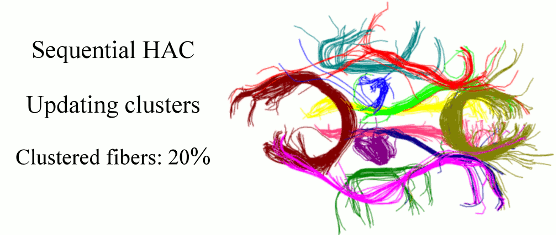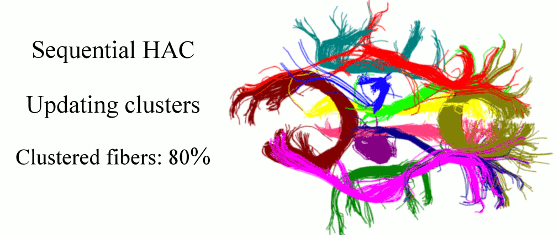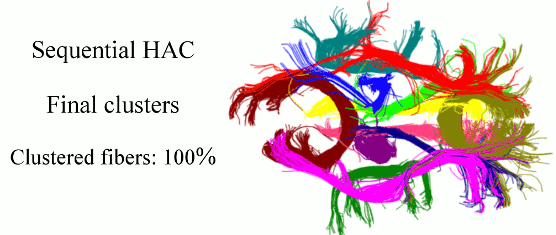
Quantitative characterization of the white matter circuitry in the human brain is paramount for mapping the structural brain connectivity. Diffusion MRI can provide information to represent this circuitry as a tractogram, i.e., 3D fiber pathways given by tractography. Following the computation of a tractogram, it is a common practice to focus on selected fiber bundles whose integrity is anticipated to be affected by development, degeneration, or disease. Therefore, automatically clustering white matter fiber pathways into anatomically distinct bundles is important for fast, reproducible, and subject-specific analysis of the brain connectivity, as well as its impairment if present. In this work, we cast this problem as clustering streams of data and develop a sequential framework to process one fiber at a time. This can be thought of as a mechanism that does fiber clustering while simultaneously accepting newly computed fibers for analysis, and as a result, alleviates the burden of computing the distances between every pair of fibers in a given tractogram. Our method represents the clusters with parametric models, performs hierarchical agglomerative clustering of relatively small number of fibers only when the parameters need to be initialized and/or updated, and assigns the labels to the following streams of data according to the current models. Experiments on phantom data evaluate the sensitivity of our method to initialization and parameter tuning, and show its advantages over alternative techniques. Experiments on real data demonstrate its efficacy and speed in clustering white matter fiber pathways into bundles that are consistent with the neuroanatomy.

