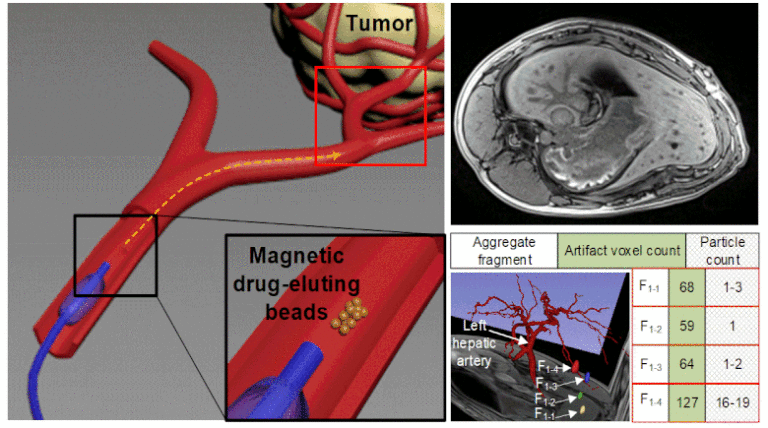
Superparamagnetic nanoparticles (SPIONs) can be combined with tumor chemoembolization agents to form magnetic drug-eluting beads (MDEBs), which are navigated magnetically in the MRI scanner through the vascular system. We aim to develop a method to accurately quantify and localize these particles and to validate the method in phantoms and swine models. To achieve this goal, MDEBs composed of poly(lactic-co-glycolic acid) and Fe3O4 SPIONs coated with C12-bisphosphonate were fabricated and then known numbers of MDEBs were injected into glass and Polyvinyl alcohol phantoms and two living swine. Susceptibility artifacts in three-dimensional (3D) volumetric interpolated breath-hold examination (VIBE) sequences of MRI were acquired. Image processing of VIBE images provided the volume relationship between the MDEBs and their artifact at different 3D T1-VIBE acquisitions and post-processing parameters. Simulated hepatic-artery embolization was performed in vivo with an MRI-conditional magnetic-injection system, using the volume relationship to locate and quantify MDEB distribution.
Individual MDEBs were spatially identified, and their artifacts quantified, showing no correlation with magnetic-field orientation or sequence bandwidth, but exhibiting a relationship with echo time and providing a linear volume relationship. Two MDEB aggregates were magnetically steered into desired liver regions while the other 19 had no steering, and 25 aggregates were injected into another swine without steering. The MDEBs were spatially identified, and the volume relationship showed accuracy in assessing the number of the MDEBs, with small errors (≤ 8.8%). The resulting volume relationship was linear, robust, and allowed for quantitative analysis of the MDEB distribution. Combining magnetic resonance angiography with digital subtraction angiography provided a more intuitive analysis of the particle distribution. We provided a proof of concept, demonstrating that T1-VIBE-based susceptibility artifacts can be used to accurately localize and quantify MDEB aggregates, which opens the door for accurately quantifying and localizing MDEBs used in different magnetic targeting techniques.