Xiaonan Zang, Rebecca Bascom, Christopher Gilbert, Jennifer Toth, and William E. Higgins, The Pennsylvania State University, USA
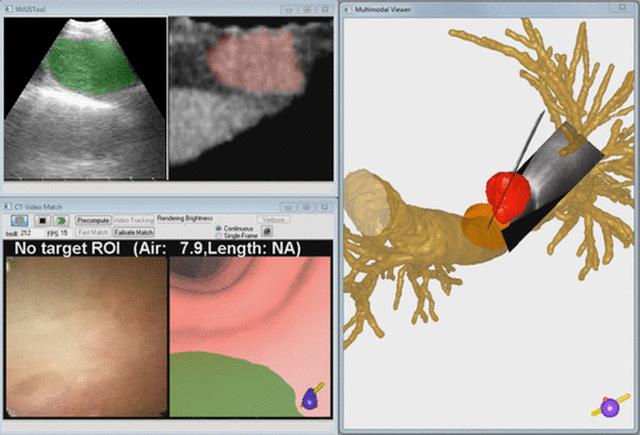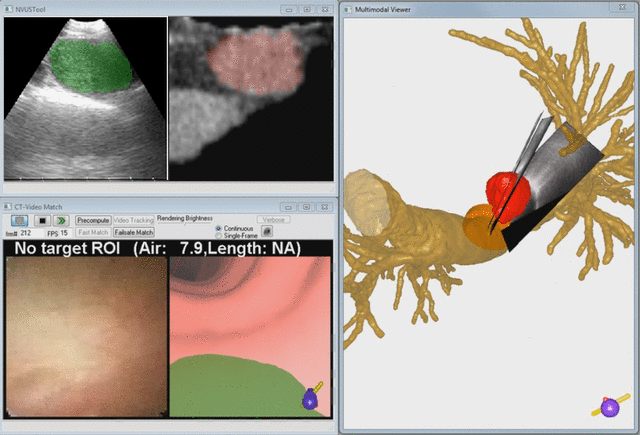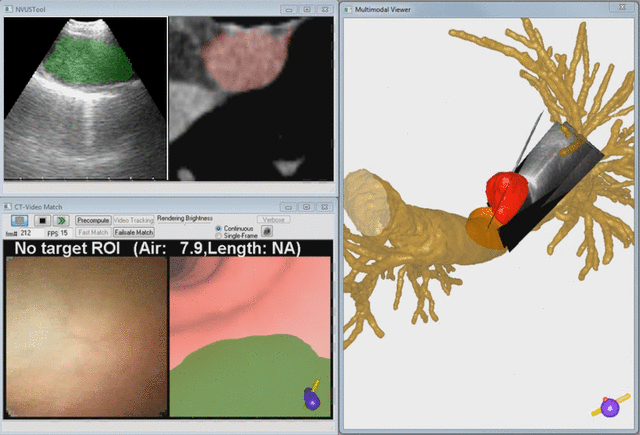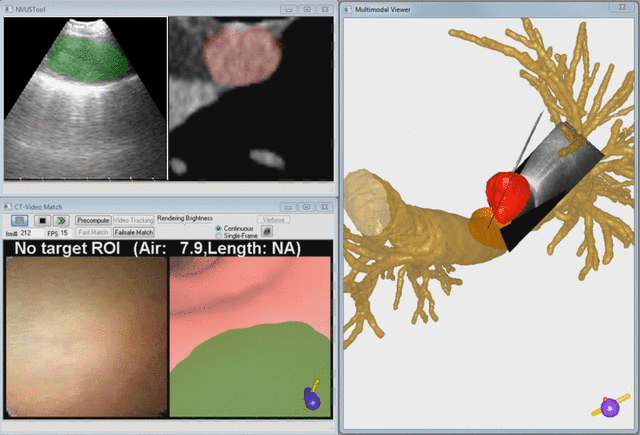
Endobronchial ultrasound (EBUS) is now commonly used for cancer-staging bronchoscopy. Unfortunately, EBUS is challenging to use, and interpreting EBUS video sequences is difficult. Other ultrasound imaging domains, hampered by related difficulties, have benefited from computer-based image-segmentation methods. Yet, so far, no such methods have been proposed for EBUS. We propose image-segmentation methods for 2D EBUS frames and 3D EBUS sequences. Our 2D method adapts the fast-marching level-set process, anisotropic diffusion, and region growing to the problem of segmenting 2D EBUS frames. Our 3D method builds upon the 2D method while also incorporating the geodesic level-set process for segmenting EBUS sequences. Tests with lung-cancer patient data showed that the methods ran fully automatically for nearly 80% of test cases. For the remaining cases, the only user-interaction required was the selection of a seed point. When compared to ground-truth segmentations, the 2D method achieved an overall Dice index = 90.0%±4.9%, while the 3D method achieved an overall Dice index = 83.9%±6.0%. In addition, the computation time (2D, 0.070 sec/frame; 3D, 0.088 sec/frame) was two orders of magnitude faster than interactive contour definition. Finally, we demonstrate the potential of the methods for EBUS localization in a multimodal image-guided bronchoscopy system.
Key words: endobronchial ultrasound, image segmentation, bronchoscopy, image-guided intervention system, lung cancer
Laboratory web site: http://www.mipl.ee.psu.edu/