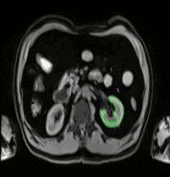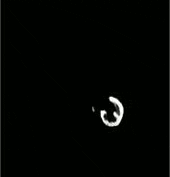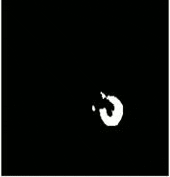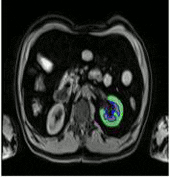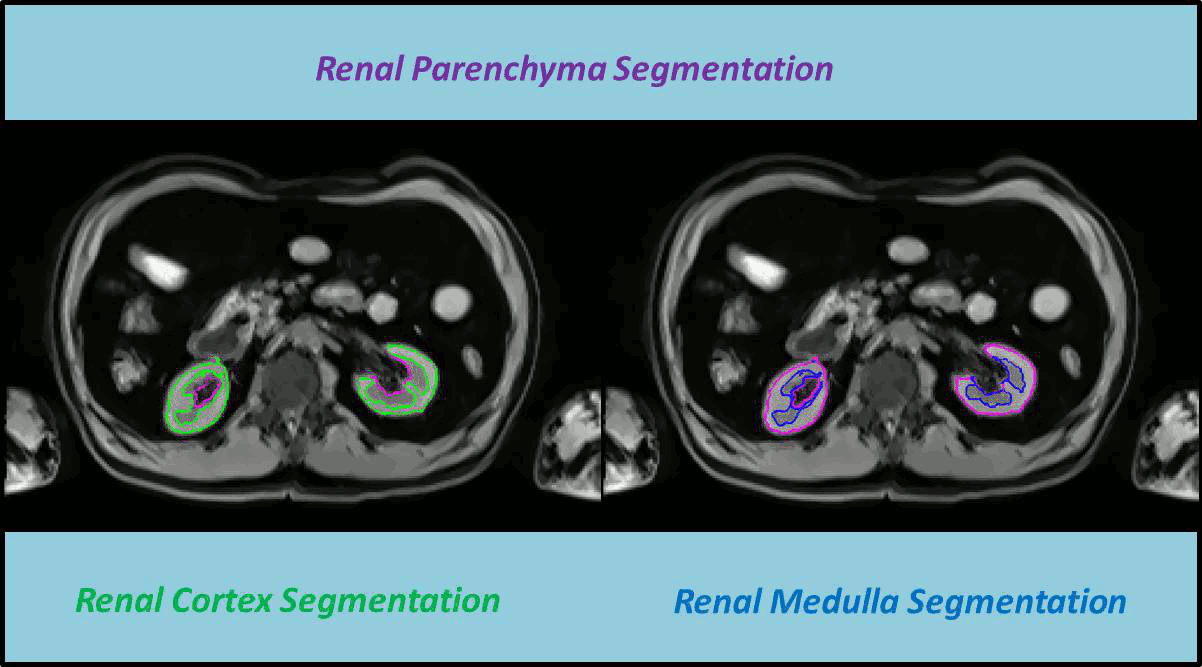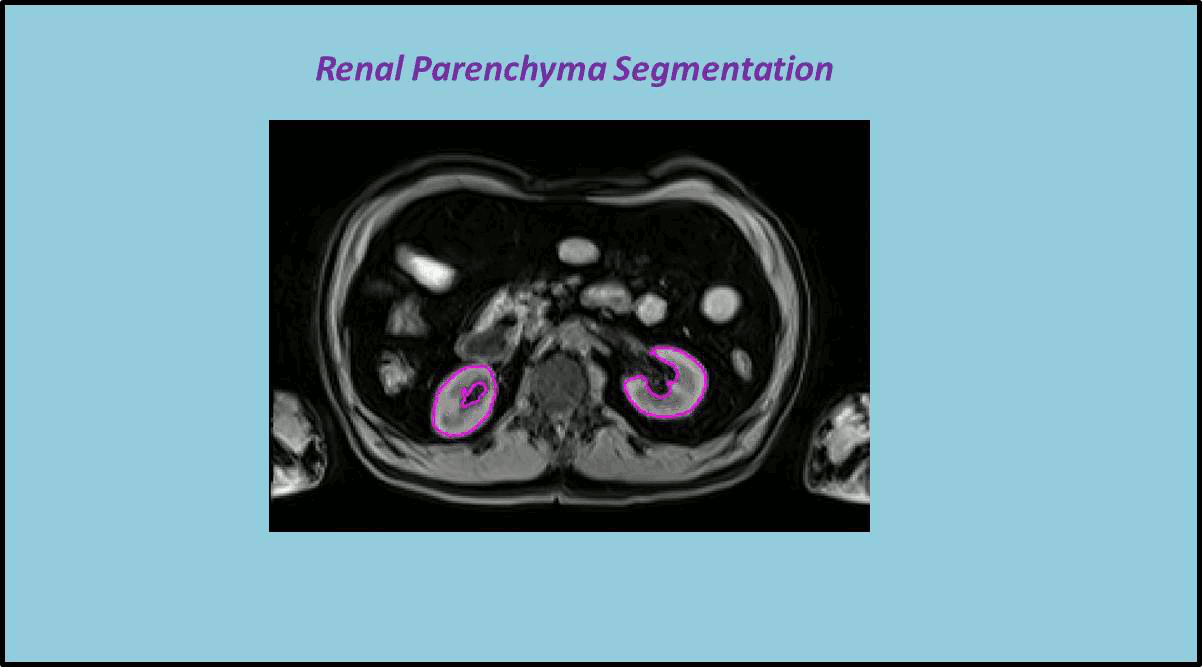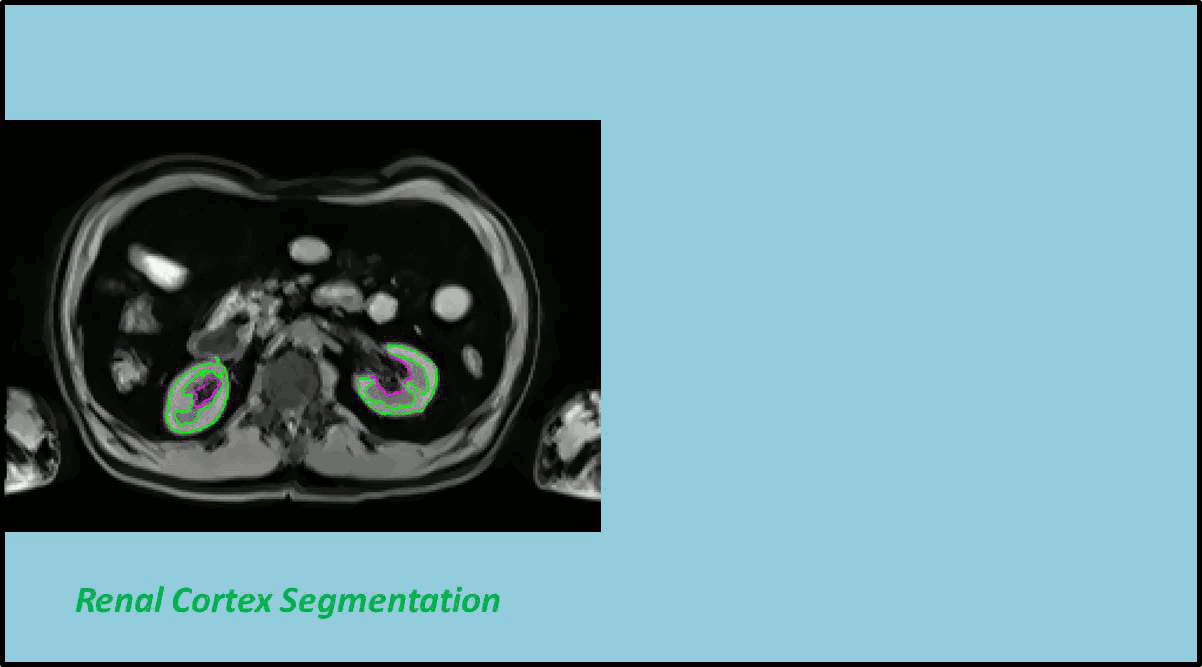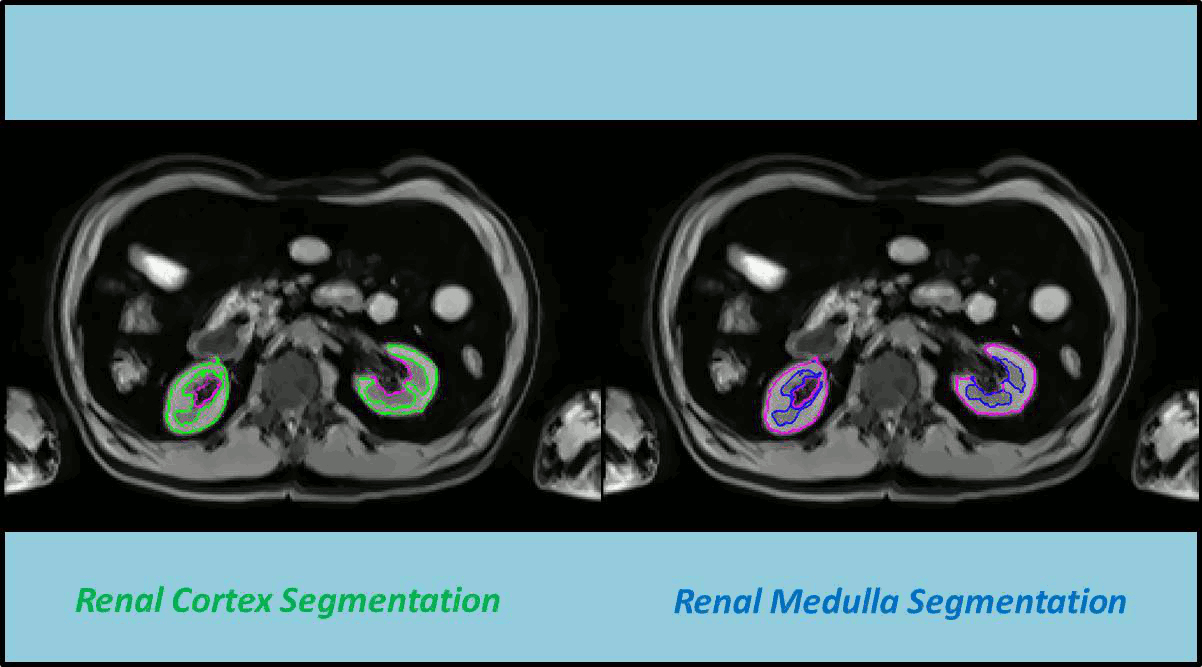
In this work a fully automated framework for renal parenchyma segmentation in native MR volume data is presented. The framework is extended by automated renal cortex and medulla segmentation to allow differentiated volumetric analysis of renal tissue types for an epidemiological study. Segmentation-based quantities like organ and tissue volumes in 3D MR image data are of increasing interest in epidemiological studies and clinical practice. Since manual segmentation is time-consuming and prone to reader variability, large-scale studies need automatic methods to perform organ segmentation. A novel strategy of subject-specific renal probability map computation is proposed, which takes inter- and intra- MR-intensity variability into account. A prior shape level set segmentation is used to segment cortex parts in transversal slices showing characteristic 2D shape that are then used to collect subject-specific samples of all available MR-weightings for subject-specific cortex probability map computation. A 3D prior cortex shape level set segmentation is applied to segment renal cortex in subject-specific cortex probability maps. 3D level set segmentation is extended by novel outer cortex edge alignment forces. After that the complete renal parenchyma is delineated in slicewise 2D convex hulls of the segmented cortex areas. This step is conducted in parenchyma probability maps using a 3D prior parenchyma shape level set segmentation. If no characteristic 2D cortex parts can be recognized, our method is designed to segment 2D characteristic parenchyma parts. 3D prior parenchyma shape level set segmentation is then applied to delineate parenchyma in parenchyma probability maps. This alternative segmentation in a single step provides the automated framework enough flexibility for application in epidemiological studies. Subject-specific probabilities for medulla and cortex tissue are applied in a fuzzy clustering technique to delineate cortex and medulla tissue inside segmented parenchyma regions. The novel subject-specific computation approach provides clearly improved tissue probability map quality than existing methods.