Ayala Greenblum, Raphael Sznitman, Pascal Fua, Paulo E. Arratia and Josué Sznitman
Technion – Israel Institute of Technology, Israel, École Polytechnique Fédérale de Lausanne, Switzerland, University of Pennsylvania, USA, Volume 61, Issue 8, Page: 2278-2289
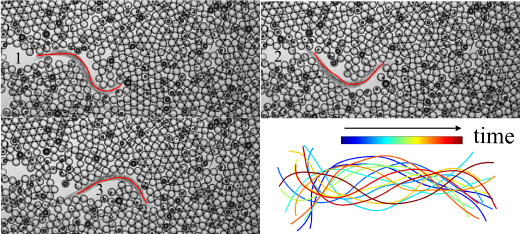
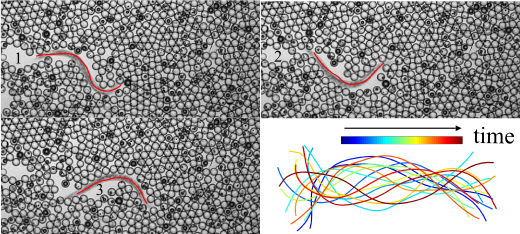
With its short life cycle, a complete knowledge of its cell lineage, a simple nervous system and a fully sequenced genome, the nematode Caenorhabditis elegans has become a ubiquitous model organism to investigate the genetics of development, neuroscience and behavior. As such, an active area of research lies in behavioral motility phenotyping of this model organism for reverse genetic approaches, where specific genes of interest are identified from whole-genome sequences. Unfortunately, linking locomotion phenotypes to genotypes remains a cumbersome and time-consuming task, requiring imaging numerous assays of nematodes and then carefully annotating hundreds, if not thousands, of image sequences manually. To alleviate this annotation burden, computer vision and machine learning tools for automated analysis have emerged over recent years in view of designing high-throughput behavioral assays. While these methods have been effective for automatic segmentation across a range of motility environments, such as crawling on substrates or swimming fluids, they remain ill-suited, if not entirely compromised, when dealing with more challenging backgrounds that are inherently dynamic and/or contain complex textures. Yet, these complex environments are increasingly relevant in view of designing artificial biomimetic micro-robots for example. To this end, we propose an automatic segmentation strategy that combines the use of intensity, texture and temporal features within a probabilistic framework to deliver coarse segmentation approximations, which are then refined using a Markov Random Field (MRF) model. We evaluated our approach on a number of well-established motility environments and show reliable segmentations for the purpose of key motility phenotypes extraction in both traditional and extremely challenging nematode essays. Results on challenging environments provide significant performance gains compared to traditional methods.
Keywords: C. elegans, motility, phenotyping, computer vision, segmentation
Website: http://biofluids.technion.ac.il