The process of rare disease identification by clinical geneticists is closely associated with the ability to correlate the phenotype of a patient with the relevant genetic syndromes. In order to perform this correlation, the phenotype has to be described in a canonical form or language. One such language is the human phenotype ontology, which defines the human phenotypes in a hierarchical form and facilitates the association between specific phenotypes and diseases. With such a structure, clinicians are able to evaluate the specific phenotypic features during the clinical evaluation process and then correlate those phenotypes to relevant diseases.
While some phenotype features are definitely objective, others also have subjective definitions. For example, the feature known as “short chin” is defined as the “decreased vertical distance from the vermilion border of the lower lip to the inferior-most point of the chin.” Phenotypes such as this are hard to define in a robust manner. In addition, the description of the phenotype using predefined terms is limited by nature, as the clinician has to decide which features are included or not. It becomes a binary decision that is made for each respective phenotype. For example, when the clinician is evaluating a patient, it is not possible to assign a portion of the feature as “short chin”—the chin is either short or not. However, in reality the relationship between the phenotypic features and correlating disease is much more fluid and features impact each other. By looking at the facial features, there may be a correlation between the length of the chin and some other features that are related to the lower face, resulting in shortened length, but not described as “short chin” and hence not considered by the clinician.
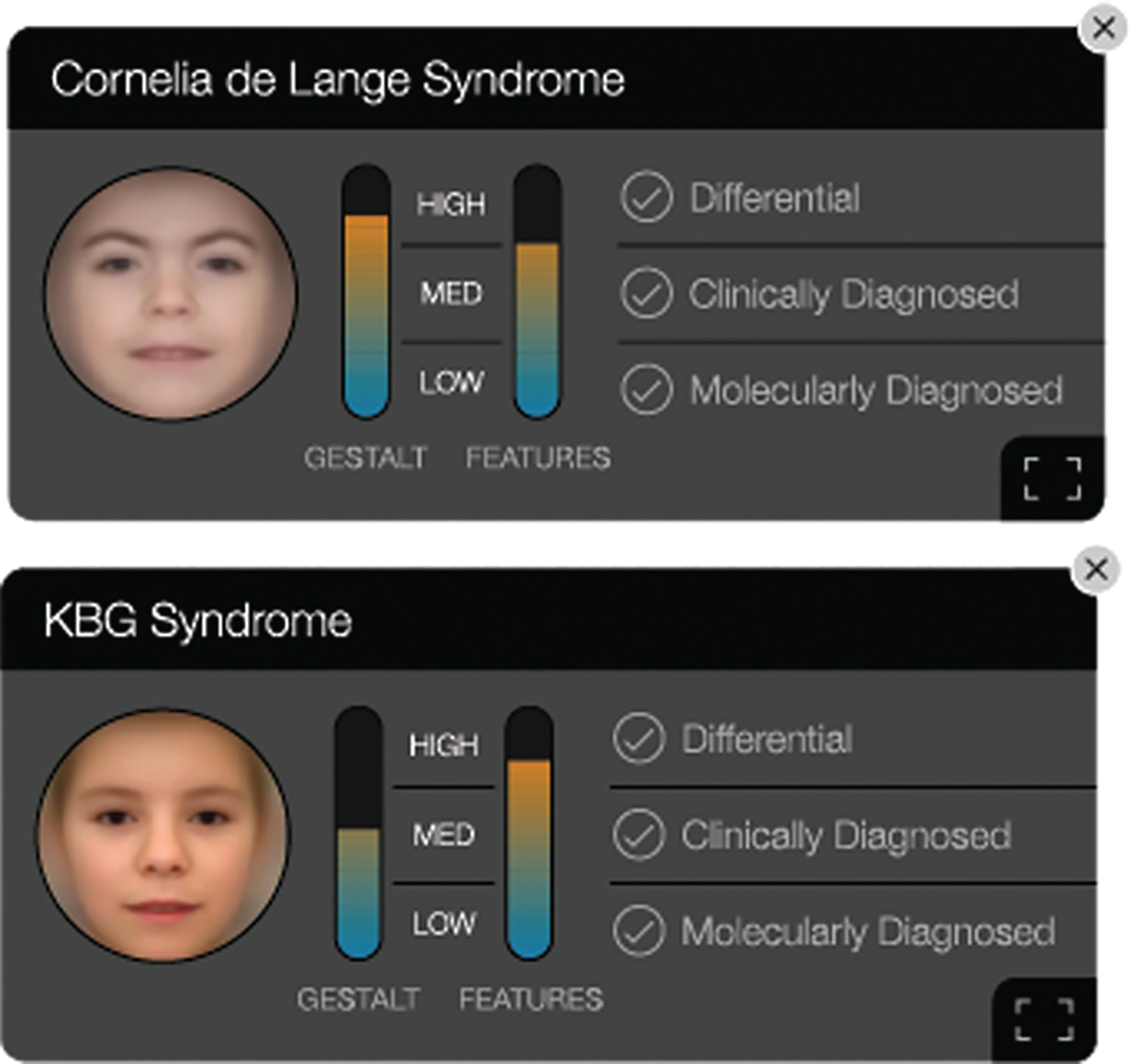
Clinical practice then has been using another method to evaluate the phenotype—the gestalt. Gestalt is defined as an organized whole that is perceived as more than the sum of its parts. In medicine, we refer to the overall representation of a specific syndrome based on the compilation of various facial phenotypes relating to that syndrome. Some syndromes have a very typical facial appearance, and clinical experts who have seen similar patients in the past will be able to accurately identify the correct disease by looking only at the face.
When talking with clinicians, they indicate facial features as the leading source of phenotypic information, with the reasoning being that in many cases it can be challenging to identify individual features, but the overall appearance, the gestalt, is more likely to be representative of a certain syndrome. In the case of rare genetic syndromes, we’ve seen a growing trend of clinicians sharing pictures and ideas with a connected community of peers regarding patients with similar presentations, and this further adds to the creation and “fine-tuning” of the gestalts.
Building an AI solution to support precision medicine
This was the main motivation in building an artificial intelligence (AI) solution within genetics—to leverage its potential to provide clinicians with an enhanced ability to correlate between the phenotype and genetic syndromes, but to also connect a community of experts to help solve medical mysteries and shorten the diagnostic odyssey that many people with rare and genetic disease face.
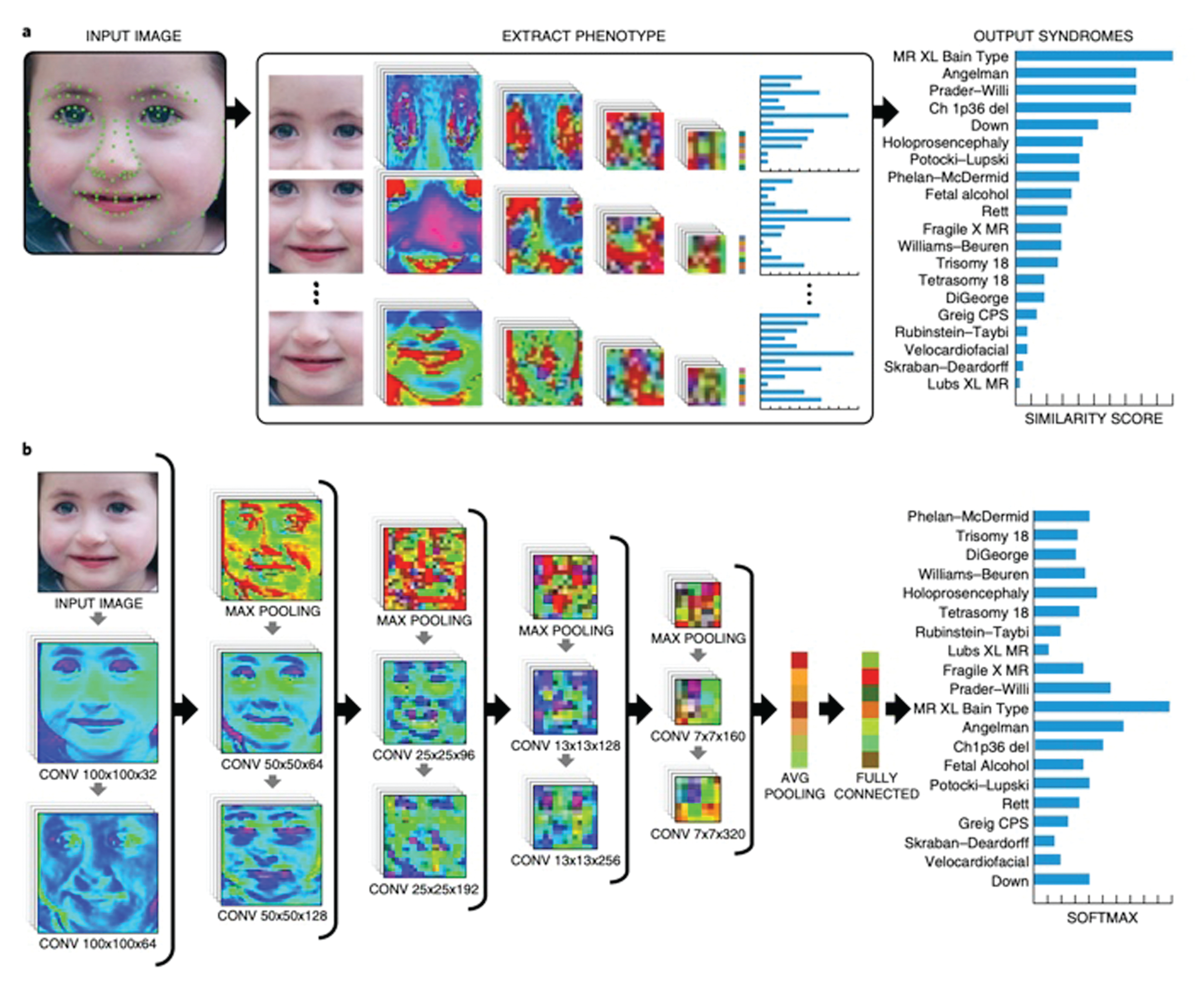
In order to build such a system, we decided to focus on facial analysis in our journey to create a leading AI solution using next-generation phenotyping (NGP).
On the technology level, the basic flow is to receive a facial image of a patient and to be able to list the top probable syndromes associated with that specific patient (Figure 1). This function is very similar to the function of facial recognition algorithms where a facial image is introduced to the system, which returns a list of the most probable gestalts that are associated with it. However, in the case of rare genetic syndromes, there are a few unique challenges.
Challenges on the road to success
One major challenge is the size of the available data. In order to build a facial recognition algorithm, one can download as many facial images as needed from the web (images of celebrities for example) and use those to train the system. If more images are needed it is not a very complex task to acquire more data. In our case, however, in the space of rare and genetic disease, most of the available data is siloed in clinics, gathered by clinicians, without the option to be shared with external parties due to privacy concerns. We established partnerships and created our flagship application, Face2Gene, to help us address this challenge.
The second major challenge is class imbalance. With facial recognition one can take as many facial images as needed per person. In our case, when the class is a syndrome, there is a natural limitation to the amount of patients that have been diagnosed with the same syndrome. For rare syndromes, this number could range from just a few patients to a few hundred. This requires our algorithm to be able to work with a very imbalanced dataset, and must be able to overcome these issues.
Another important challenge is the ethnic diversity of the databases. When aggregating knowledge from clinicians, we noticed that there is an internal bias of ethnicity that is closely associated with certain syndromes, some more than others. Lastly, we aspire to support thousands of syndromes with this type of algorithm, but as the amount of supported syndromes increases, it becomes more challenging to keep overall performance at a consistently high level, which introduces yet another challenge in building an AI system such as this.
Our approach to this challenge was to establish an ecosystem where there exists a virtuous cycle of value that feeds itself and allows growth by its nature—Face2Gene serves clinicians and provides services that improve both routine work in the clinic and also facilitates research activities. This technology is providing value to our network of users, which by using the system provides data that is then used to further train the system and provide enhanced technological ability and more value to those same users. This is the resulting cycle that facilitates a win–win for the tech developers, health care providers, and patients.
To illustrate this point further, users are able to use Face2Gene CLINIC while meeting patients in person. The image of the patient will be uploaded to the system, the clinician will instantly receive the NGP analysis and will be able to provide better clinical care by either helping to confirm a diagnosis or determine a targeted test option. Using the selections the clinician added to the case, and potentially the molecular tests that were performed based off of the applications recommendation, the system will be able to adjust to errors and be trained to more accurately and efficiently recognize more syndromes for future patients.
The process of research and development for such AI-based products includes a few special considerations, such as what is the benchmark that the system is evaluated on, and how likely it is to reflect the real cases that will be seen outside of product testing.

In this case we used blind testing of real, deidentified cases from users that had previously been uploaded in the system. As we maintain the highest privacy and security standards, developers are unable to access user data.
Lastly, we needed to determine how the recommendations from the algorithm should be explained to the user, and even before that, how we can foster trust among clinical users to integrate this technology into their workflow and ensure the results are meeting their needs. For this, we introduced heatmap visualization that highlights the areas in the face which had the maximum correlation to each syndrome respectively (Figure 2). Clinicians look at these heatmap representations and are able to understand why the algorithm ranked a syndrome the way it did.
Another powerful tool we use to depict the algorithm’s reasoning is the composite image, where we average the training facial images and create a gestalt that highlights the appearance of a specific syndrome (Figure 3).
The future of AI-assisted health care
In today’s rare and genetic disease landscape, great strides are being made by genetic testing labs such as Illumina and PerkinElmer Genomics that improve targeted testing and lowering costs to do so, and more broadly utilizing AI solutions within the genomics field as a whole. Even as advancements in technology continue to be made and serve to increasingly foster connections and interactions between humans, some tasks in practicing medicine must still be left solely to humans.
When asked about his experience with AI-based technologies like Face2Gene, Dr. Abdul-Rahman, director of genetics at Munroe-Meyer Institute, concluded, “It helped us get a diagnosis without me ever laying a hand on this patient, who lives literally four or five hundred miles away, and the diagnosis was already suspected before they walked through the door because of that intake process.”
Tailoring a solution that combines the components of genomics and AI-based phenotyping is not an easy task and it depends on the ability of stakeholders from many disciplines to work together, share data, and collaborate on research and development. To achieve this, integrity of the data, ethical and privacy policies, and trust in workflow should be established, which requires an open dialogue between all parties involved and a fast-paced framework to allow developers to move quickly in building these tools.



