On a video screen, against a black backdrop, 15 spherical blue-green cells vibrate with a quiet energy. Slowly at first, then faster, they begin to roil and roll. Within the confines of their round membrane cases, they divide, becoming two, three, four cells, then those, in turn, divide to become eight. One splits into two, then pauses, struggling to catch up and spinning off pieces of cellular detritus as it does. Near the top, another, by now many cells rich, hollows out and expands, contracts, expands, contracts. It falls in upon itself and then hatches, pouring out from its shell and ballooning to the side.
The process is beautiful and messy—and, in a collective sort of way, it is us. These are 15 human embryos, photographed every five minutes over the course of the first six days of their development. This particular film comes out of Stanford University in a study led by renowned stem cell researcher Renee Reijo Pera. It began in the mid-2000s, when Reijo Pera, hard at work mapping the molecular machinery behind early human development, took a look at Stanford’s Photonics and Mechanical Engineering Departments and had a realization. “Boy, there are a lot of people here who know how to make microscopes,” Reijo Pera, now vice president for research at Montana State University in Bozeman, remembers thinking. “Why don’t we link that map to visuals?”
From that collaboration, a sleek new visual system took shape, harnessing dark field imaging, heavy-duty computer algorithms, and miniaturized microscopes and camera equipment that could fit inside a standard clinical incubator. And for Reijo Pera, the results were astonishing. As she peered through her microscopes and films, she was struck not by the chaos of it all—the rolling, writhing embryos, the cellular fragments—but the regularity hidden within. Despite the overall tumult, the embryos that went on to become blastocysts moved discernibly differently than their failed peers, and similar to one another. They were smooth and steady; it took them about 14.3 minutes to complete the final stages of the first division into two cells, 11.1 hours to go from two to three cells, and another hour or less to reach four cells. Indeed, so reliable were these numbers that they could fit into an algorithm and be used to forecast the fate of other embryos—which is exactly what Reijo Pera did. Her team created a fully automated computer analytics system that could go from fertilization to blastocyst forecast in two days flat, with a better than 93% sensitivity.
In many areas of science, time-lapse imaging, or turning photographic series into films is an old idea, but in the study of human embryology, it has really just arrived in the last five years. Yet in those five years, it has already opened windows into humanity’s earliest days that researchers had barely considered only a few years earlier. By now, a new science is underway, forming around the ebbs and flows of our embryonic development and what they may reveal about our futures and our genetics. It is a teaching tool, a basic research tool, clinical guide, and the cause of at least a few debates about the universality of human patterns of development.
“It’s been very, very fascinating to me that we spent decades crunching up cells, making RNA and DNA out of them, and thinking we knew what was happening,” Reijo Pera states. Now, “the idea of looking is something I’ve become fond of. I love it because it seems like it was a bit on the old-fashioned side—Linnaeus and Aristotle, they walked around and looked at things and named them. And now we’re looking again, and there’s this kind of beauty in the science.”
Changing the Game
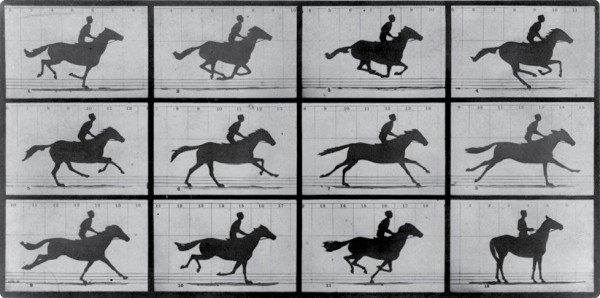
Time-lapse imaging via various means has existed since the late 1800s, when its founding father, English photographer Eadweard J. Muybridge developed the technique using tripwires and multiple cameras as a way to study physical movement in animals and humans (as well as to settle a famous debate about whether all four of a running horse’s hooves are ever off the ground all at once—they are; see Figure 1). Following his example, scientists seized on the tool and, over the course of the next century, adapted it for such things as the X-ray cinematography of organs, microscopy of blood-borne parasites and cancer cells, and even more recently, electron microscopy to capture the minutia of forming brain circuits.
Primordial rabbits were the first embryos to fall under the lens of a time-lapse device, as part of a 1929 Science publication. Again, researchers took note, and various species quickly followed suit. Today, time-lapse microscopy serves as a crucial element in the modern study of embryogenesis, from the chicken and mouse to the fruit fly and zebrafish.
But humans are a different matter altogether. Due to the many safety and ethical complications that surround human embryo work—not to mention fact that the embryos themselves were entirely inaccessible until 1978 with the advent of the first successful in vitro fertilization (IVF) treatment—time-lapse work has been scarce in homo sapiens. A few papers did appear now and then in the literature, starting around the late 1990s. One of the first landmark attempts painstakingly imaged 50 fertilized eggs one at a time using a video camera, microscope, switching box to automate a light on the scope, and an environmentally sealed chamber. But such studies were difficult and limited, and the work never gained in popularity.
That is, until around the end of the last decade. In 2010, Reijo Pera and her team published their time-lapse findings in a seminal Nature Biotechnology paper (Figure 2); two years earlier, they had already founded a California start-up called Auxogyn to begin commercializing the scope-and-algorithm setup, which they dubbed Eeva (Early Embryo Viability Assessment). In one of those curious moments in science when the same idea suddenly and independently appears from multiple sites, two other major commercial-grade embryo time-lapse monitoring systems appeared. Out of Hungary, veterinary researchers on a mission to evaluate their cloning and freezing research had created a similarly compact system—Primo Vision—reminiscent of Eeva but without the automation. In Denmark, meanwhile, microsensor experts from the Aarhus University spinoff Unisense realized that scientists were showing far more interest in the data rolling off the time-lapse microscope that they had tacked into an incubator originally designed to assess embryo oxygen use, and they reworked it into an all-in-one time-lapse incubator now known as the EmbryoScope.
Slowly at first, but with increasing speed, the use of and investigations with these devices have gained steam. Data are coming in regularly from labs in the United Kingdom, United States, Spain, France, Denmark, Norway, and beyond. Granted, research articles on the topic still number in the dozens rather than hundreds, and only a fraction of the world’s roughly 5,000 clinics have put these tools into use—but those numbers are growing.
Part of the reason for these devices’ current success is their relative safety and practicality. They either are or fit into clinical-grade incubators. They boast large embryo capacities—72 at a time for the EmbryoScope—and sophisticated software that feeds photos to external computers and servers and helps researchers mark time points and analyze data. They also all arrived at the right time, in the midst of an active research period by the assisted reproduction field into alternatives from morphology—the gold-standard for embryo assessment in IVF, in which embryos are briefly removed from culture and examined under a microscope—that would help improve the field’s famously tepid success rates. (See also “The Birds, the Bees, and Technology” and “Babies from Skin Cells” by Leslie Mertz in the January/February 2015 issue of IEEE Pulse.)
“We were all trying to go away from morphology by going into investigations on media culture, on proteins, on amino acids, on nutrients, on prior rate glucose,” explains Markus Montag, a longtime embryologist and founder of the reproductive medicine consulting firm Ilabcomm in St. Augustin, Germany. “And then came these devices which simply only took pictures, and pictures are only morphology. But all of a sudden this morphology was related to the time of development—meaning that, for the first time, embryologists were able to access cell cycles.”
Crisis of Universality
Since this time-lapse renaissance began, competitor devices have emerged, although Eeva and the EmbryoScope, even more than Primo Vision, remain the two dominant systems. Crucially, the EmbryoScope does not have Auxogyn’s automated algorithm—instead, users observe the embryo activities directly, annotate key events, and devise their own predictive algorithms. It requires a steeper learning curve but allows for more freedom and open-ended research.
Nina Desai directs the laboratory at the Cleveland Clinic’s fertility center in Ohio. She first encountered modern time-lapse devices at a reproductive research conference in 2011, then bought one—an EmbryoScope—for her lab in 2012 (Figure 3). By the end of the year, she had bought three more. “It’s an amazing technology,” she says. “I’ve been blown away by some of the things I’ve seen.”
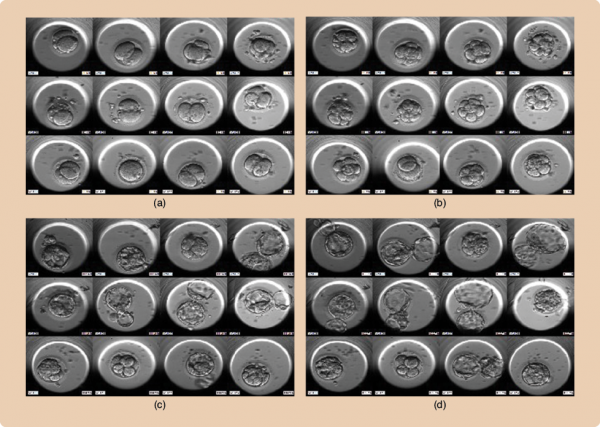
When Desai set up her first device, she brainstormed a host of potential embryo health predictors then kicked off an exploratory retrospective trial with patients in her clinic. Over the course of time, she winnowed those parameters down to a few that were associated with an embryo’s development into a blastocyst and several others—not always overlapping—that correlated with embryos that successfully implanted into their mothers’ uterus to begin pregnancy (Figure 4). These included markers like the rate at which the fertilized eggs reached two, four, and five cells, and how synchronously they did so. A little more than a quarter of the embryos Desai saw, generally the least successful, also showed bizarre anomalies, like going from four cells back to three—”reverse cleavage”—or leaping ahead, from one to three cells or from two to five—”direct cleavage.”
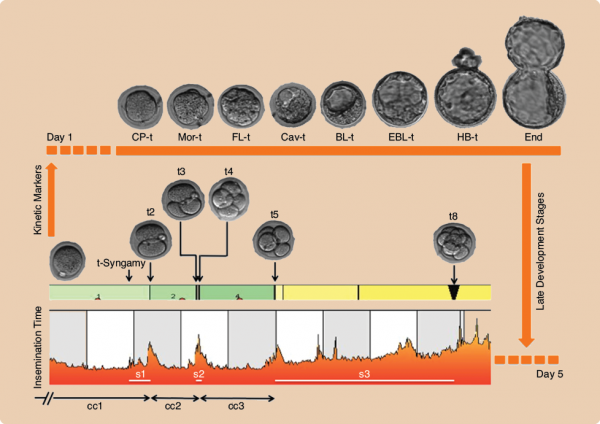
In the language of the field, gross missteps like reverse and direct cleavage are roundly considered “deselection parameters.” They are one of two main themes emerging from the building data. They are also the type of anomaly that showcases time-lapse’s appeal, since such parameters are not only widely acknowledged trouble signs, but also invisible with standard once-a-day morphology assessments. “You get deceived into thinking you’ve got a four-cell embryo on day two and you think it’s fine,” Desai says. “But in reality, it went from one cell to three cells and then it became a four-cell—and that embryo, from the data I’ve seen, has very little chance.”
The second theme consists of the finer cell interactions and timings—how long it takes to go to four cells, how synchronously, and so on. These are the “morphokinetics,” and they are far subtler and more controversial, and a lack of consensus in the literature over predictive variables has become typical.
Micronucleation 2 and 4 Cell
Part of the problem is that the field is still finding its feet. Particularly with more manual devices like the EmbryoScope, investigators have not come to an agreement on how to define and annotate embryonic events. Mixed results may also be due, believes Simon Fishel, managing director of the U.K.-based CARE Fertility line of clinics, to inevitable lab-to-lab differences.
“Embryos are clearly very sensitive to the environment in which they’re in,” he explains. “And if you’re using a different culture medium than me, or I’m using a different type of time-lapse system, then maybe the whole environment is different for the whole five-day period, temperature, gas fluctuations—all those things—and so it may well be that an algorithm is right in one lab but not in another.” Fishel’s team recently tried an embryo selection model originally developed in Valencia, Spain, but found that in his clinics, it failed to predict a successful pregnancy.
Multinucleated Embryos
Similar issues can arise even between branches of the same clinic. At Norway’s Klinikk Hausken, IVF Lab Manager Shabana Sayed spent several years developing a tiered algorithm for variables she identified using EmbryoScopes in her labs. But the algorithm that her team developed in its Haugesund branch, Sayed says, did not apply exactly to its Bergen one. “It’s mostly the same set of doctors because it’s a satellite place,” she says. “But it’s different hands that are working with the embryos, and everything matters. Now we’ve had to develop a separate algorithm for them.” While the two algorithms are similar, she says, they are not identical.
All this contradicts Auxogyn’s thesis—that is, that there is a universally applicable set of variables reliable enough to be automated in a computer program and applied in any lab right out of the box. It is a thesis that has been further bolstered by a recent multicenter trial reporting that their Eeva model, albeit further simplified, could be readily standardized and used to improve embryologists’ embryo selection across five different California fertility clinics, despite different tools, clinicians, and environments. Yet, on the other hand, a different study that retrospectively applied those Auxogyn parameters to data from seven clinics in three countries found that while the model would have increased implantation rates by 30%, it would also have rejected a substantial group of embryos that did implant.
It’s a controversial point, says Montag, speaking of the ongoing back and forth about the universality (or not) of human developmental patterns. He suspects that certain universal themes will emerge in the predictive data—for instance, it inevitably takes time for a cell to fully replicate its DNA, and if a cell division occurs too quickly for that to have happened, then the embryos simply would not be adequately equipped no matter the environment. But he believes that there will also be error bars on that time interval and that it will be up to the laboratory in question to find the best algorithmic fit for its situation. “And people are doing that,” he says, “It’s already a tradition, at every conference, [that] you hear somebody talking about a published algorithm and how they fit to their lab or not. And you can see mostly [the data] don’t fit, but once they start to adapt it a little bit, they get something out of it.”
Genes at a Glance?
The most obvious motivation behind much of this work is in the IVF clinic. Caregivers hope that time-lapse imaging will provide a way of gauging an embryo’s odds that is less invasive than a genetic biopsy, but more accurate than morphology. So far, the data are generally promising, if still accumulating.
But the potential applications go far beyond that. For instance, Desai has discovered her EmbryoScope can be an unexpected boon for embryologist training. Rather than learning what embryos should look like with fleeting glances once a day, her new staff now sit with senior embryologists as they grade the embryos for later IVF transfer, taking the time to fully understand what it is they’re watching. “This detailed daily grading and critique by senior embryologists serves to expedite the training process such that within six months we could feel confident in assessments performed by our junior staff,” explains Desai.
The more precise and detailed information that time-lapse allows may also help troubleshoot practices. For instance, culture medium—the liquid bathing embryos—remains a hugely controversial topic: some clinics use a single medium throughout the incubation period; others change it up sequentially. Some formulations contain more or fewer ingredients, and some are proprietary information not fully available to the labs that use them. In 2012, researchers led by Haydar Ciray, then at the Fulya IVF Clinic in Turkey, now at the University of Leeds in the United Kingdom, reported kinetic differences in embryos cultured with a single versus a sequential medium, beginning with the very first cycle of divisions, possibly, the authors speculated, due to amino acid content in the different media. What does it mean? That is not clear yet, but research elsewhere in the field has begun to suggest that stresses from embryo culturing may produce epigenetic changes, the extent and risks of which are still unknown. Time-lapse, particularly in conjunction with genetics and epigenetic work, may be able to help pinpoint where and how such changes occurs.
The groundwork for that type of molecular work is underway as well, some of it from within Reijo Pera’s own lab, where she has held true to her original ambition to map molecular events. As part of the 2010 Nature Biotechnology paper, for example, her team also reported that abnormal embryos showed chaotic gene expression profiles, while those that developed normally—kinetically speaking—displayed a sequence of four unique gene patterns, including one from the egg, and three more associated with the early embryo’s various stages of development.
More recently, she has also begun exploring time-lapse and epigenetics in both humans and mice. Elsewhere, at the University of Melbourne, in Australia, researchers have published the first study so far correlating morphokinetics with embryo metabolism—how much oxygen and amino acids an embryo burns through and produces, for instance. And meanwhile, multiple labs are industriously chipping away at the links between morphokinetics with general chromosomal health. All of this is preliminary, but it may be the beginning of a new, hands-off way to look at an embryo and know what is happening at the molecular level to drive the behaviors on screen.
This work is only beginning—there are still so many parameters and biological correlates left to explore, perhaps even embryo and cell behaviors or patterns that the naked eye might not be able to see, but that computer algorithms and vision will be able to catch and make sense of. “We’ve only touched the tip, the easy things to look at,” Desai says. “But now if you sit there and really analyze all the patterns, and all the spatial orientations, the cells as they divide,” she explains, the potential for research expands far further.