With the advent of low-cost computing platforms, such as Arduino and Raspberry Pi, it has become clear that lowering the cost barrier and shortening the learning curve, with the backing of a motivated community, would play a transformational role in the way people learn, experiment, and create imaginative solutions to outstanding problems that can benefit from embedded systems.
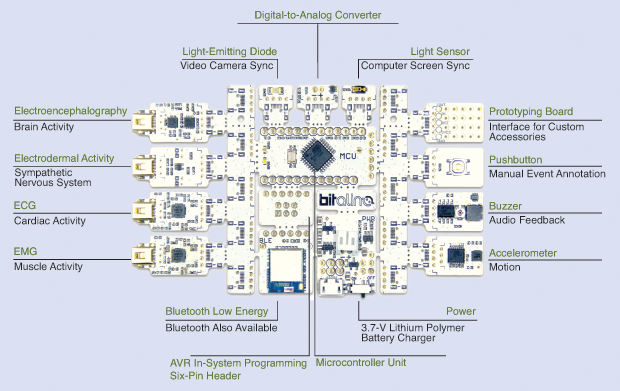
Within biomedical engineering, physiological sensing is following a similar path, with the added value of low-cost and open-source tools that policymakers and practitioners across different sectors as well as the general public find increasingly useful. This idea has been further reinforced with the recent recognition by the European Commission’s Directorate General for Communications Networks, Content, and Technology (commonly known as DG-CONNECT) of the biomedical tool kit BITalino as the best “Industrial and Enabling Tech” of 2017 within the scope of the organization’s Innovation Radar Prize (Figures 1 and 2).
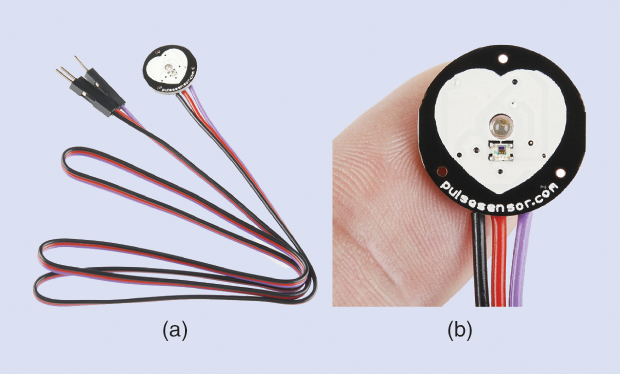
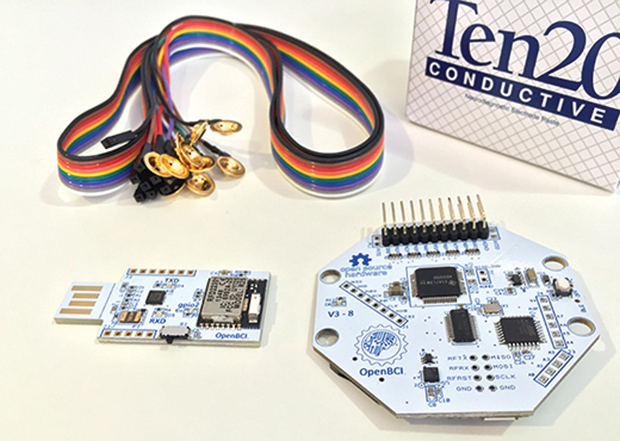
Today, there is a growing number of other readily available resources in this area (i.e., specifically designed with biomedical applications in mind). Examples range from simple sensors—such as the PulseSensor for photoplethysmography (PPG), shown in Figure 3, or the MyoWare for electromyography (EMG)—to the OpenBCI system, which supports the design of brain–computer interfaces (Figures 4 and 5), and hybrid solutions, such as the Libelium e-Health Sensors shield for the Arduino and Raspberry Pi. There’s even the case of Tim Omer, a diabetic patient who took upon himself the challenge of developing a do-it-yourself (DIY) artificial pancreas.
This trend opens an array of novel opportunities for biomedical engineers, especially due to the spillovers into areas where physiological sensing is not yet traditional, such as human–computer interaction, the performing arts, and self-quantification, among many others. Specific domain knowledge is required, which biomedical engineers are very well positioned to readily provide. Moving from raw data to meaningful information in a user-friendly way still poses multiple barriers for new entrants in fields related to sensor usage, signal interpretation, normative data analysis, and related topics.
Even within the biomedical field, individuals and/or teams working with physiological sensing are often forced to develop their own tools, at great cost of time and other resources. Open source has the potential to unify such efforts, make them more effective, and create added value synergies across knowledge areas. An interesting demonstration of this dynamic is the work by Sebastian Madgwick [1], who, in 2009, developed an open-source inertial measurement unit and the attitude and heading reference system sensor fusion algorithm as part of his Ph.D. degree research at the University of Bristol, United Kingdom. Among other applications, the work presented in [1] has been adopted by researchers at the Institut de Recherche et de Coordination Acoustique/Musique and integrated into the low-cost R-IoT hardware platform (an Internet of Things hardware system) [2], now empowering hundreds of artists worldwide to create novel motion-based musical instruments and body-based media interaction projects.
The process of opening physiological sensing to the world started almost two decades ago—with the PhysioNet family of resources [3] being perhaps the most well-known initiative having such a focus. However, especially due to the latest advances toward lowering hardware costs and making applications more DIY-friendly, this area is experiencing revamped and intensified efforts around the globe. The widespread access to hardware has brought back to the forefront challenges related to the overall context of physiological sensing outside of the lab; for example, issues such as effective and efficient ways to deal with noisy data collected in highly dynamic use cases (e.g., while running) and the usability of such sensing devices for everyday life (e.g., sensing brain activity) have not yet been sufficiently addressed.

Other core challenges are related to making real-time and offline feature extraction more off the shelf. Even simple operations—such as deriving the linear envelope from raw EMG data to drive a controllable output on an Arduino or reliably computing heart rate from PPG and electrocardiography (ECG) data on an Android app—are quite challenging for the multiple user profiles currently gaining access to physiological sensing tools. At a higher abstraction level, there are also challenges related to providing turnkey solutions for pattern recognition tasks useful in end-user application (e.g., gesture recognition from motion or multimodal physiological data). Recent examples include a biosignal-specific processing pipeline for MATLAB [4], a biocybernetic loop engine for video game creation [5], and a toolbox for biosignal processing in Python [6].
In essence, as the challenges related to the availability of suitable hardware are mostly addressed, the most pressing needs are increasingly related to making the software layers more off the shelf and “human friendly.” This needs to be achieved mainly by providing tools that are accessible to audiences that can currently range from preschoolers with no knowledge of the field to professionals and researchers alike. No one is better poised than the biomedical engineering community to spearhead such an endeavor.
References
- S. Madgwick, A. Harrison, and R. Vaidyanathan, “Estimation of IMU and MARG orientation using a gradient descent algorithm,” in Proc. 2011 IEEE Int. Conf. Rehabilitation Robotics (ICORR), pp. 1–7.
- E. Flety. (2017, Mar.–July). A comprehensive guide to using, programming & flashing the R-IoT WiFi sensor module. Prototypes & Engineering Team (PIP)-IRCAM. France. [Online].
- A. Goldberger, L. Amaral, L. Glass, J. Hausdorff, P. Ivanov, R. Mark, J. Mietus, G. Moody, C. Peng, and H. Stanley “PhysioBank, PhysioToolkit, and PhysioNet: Components of a new research resource for complex physiologic signals,” Circulation, vol. 101, no. 23, pp. 215–220, 2000.
- A. Nouhi. (2017, Mar. 25). A GUI on biosignal-specific processing pipeline. MathWorks. [Online].
- NeuroRehabLab. (2018). Biocybernetic loop engine. [Online].
- GitHub. (2018). Biosignal process in Python. [Online].