If CSI and those other police procedural TV shows are to be believed, criminals don’t have a chance. A finger smudge on a light switch, a flake of skin, or a sweat-stained fiber is all the information an investigator needs to positively identify the perpetrator and put him or her behind bars.
“When we first saw CSI 15 years ago or so, we said that’s ridiculous; it doesn’t happen that way. But I’ve been in this field all my life, and I have to say I’ve been blown away, particularly over the last five years, in how far things have come. I hate to say it, but we actually are now getting kind of CSI-like,” says Timothy Palmbach (Figure 1), who spent 22 years with the Connecticut State Police as a detective in the major crimes unit, as well as also a specialist in crime scene analysis and forensic science. Palmbach is now a private forensics consultant, as well as professor and chair of forensic science at the University of New Haven (UNH) in Connecticut, which launched the nation’s first master’s degree in forensics technology in 2015 [1].

It was the fast-moving technological advances in firearms, bloodstain, and other forensic analyses, as well as crime scene reconstruction, that led UNH to pursue the idea of a graduate program in forensics technology, Palmbach says. “There was a need to address the reality of the crime scene world, where more and more of these technologies—whether it be a Raman spectrophotometer or DNA analyzer—are becoming portable. To better assist investigative needs and improve crime scene analysis, we needed to move from a lab-based model to a field-based model.”
At the same time, law enforcement agencies had begun to see the necessity of specially trained individuals who understand the capabilities and limitations of the technologies and can put them to best use. UNH spent three years on curriculum development and added four new faculty members and about US$1 million worth of high-tech equipment to get the program going. The first class in its forensics technology master of science program graduated in May 2017.
The Changing Face of Forensics
Some of the most important improvements in law-enforcement investigations fall under the heading of crime scene documentation, according to Palmbach. This goes beyond traditional video and drawings and adds the capabilities of alternate light-source-based tools for enhancing bloodstains, gunshot residues, and fingerprints; drones for overhead views; and three-dimensional scanning of the scene. “You can virtually recreate every attribute, every aspect of that entire scene and know every dimension and relative position, and you can change the perspective and the modeling,” Palmbach explains.
The portability of instruments is also key, he asserts. “You’re basically taking the same, big, expensive, highly complex, complicated- to-use, sophisticated instruments from the lab and bringing them down into a portable mode. And you’re also bringing the library of searchable databases out to the field.” As an example, he points to the discovery of a bit of white powder that may be a biological threat. “In the past, we’d have to wait days to get the results and find out what it was, so we’d have to assume the worst, go through all the precautions, do the decontaminations, which could cost perhaps hundreds of thousands of dollars. Today, with a very simple procedure, a field analyst only needs about ten minutes to conduct an analysis, do an on-board database search, and determine whether that’s some other kind of known chemical or biopathogen.”
Rapid in-field analysis of deoxyribonucleic acid (DNA) is also having an impact on forensics investigations. In fact, Palmbach is collaborating on a project to help prevent criminal activities, such as illegal baby transfers or human trafficking, at a very heavily used border crossing in Europe. “Here, we might want to test an alleged family unit that is coming across the border with a baby but either they don’t have the correct documentation or you’re concerned about the validity of the paperwork,” he says. “In that case, we can actually do a DNA analysis of that whole family by taking a buccal swab, and, with portable rapid DNA analysis, we can get an indication in a approximately 80 minutes and with potentially a 99.5% correlation to determine whether that baby is actually theirs.”
He sees such identification capabilities and biometrics continuing to improve, but there’s still a long way to go. “The reality is that it’s still a lot harder than those TV shows suggest to do forensics work. We have a lot more glitches and things go wrong a lot more often, but we are truly getting closer to those kinds of technology becoming a reality.”
Mixing It Up
One area of forensic evidence that has always been problematic is mixed DNA samples. For instance, police may have video of a bank robber opening the door to enter the bank, so they know his DNA is on the door handle. Unfortunately, the perpetrator isn’t the only person who touched that door handle. The first step in separating out those individual DNA profiles is to determine just how many contributors are in a sample.
One way to do that is through a new technology announced in March 2017 by Michael Marciano and Jonathan Adelman, two research assistant professors with the Forensic and National Security Sciences Institute at Syracuse University’s College of Arts and Science in New York (Figure 2). Combining Marciano’s background as a molecular biologist and forensic casework analyst and Adelman’s expertise in computer science and statistics, the two researchers developed the Probabilistic Assessment for Contributor Estimate (PACE), which uses machine learning to quickly determine the number of contributors in a sample.
PACE improves on currently used methods: notably, the maximum allele count (MAC), which becomes less accurate as contributor numbers increase, and the Markov chain Monte Carlo (MCMC), which, while very accurate, is computationally intensive and time-consuming, Adelman says. PACE, in contrast, is both accurate and quick. In 400 sample comparisons between MAC and PACE [2], both could correctly count the number of contributors every time in a single-source sample; however, PACE beat MAC in three-contributor samples, with 97% accuracy versus 89%, respectively, and PACE also outclassed MAC in four-source samples, with an accuracy of 96% compared to the complete failure of the MAC method. Comparisons of PACE versus MCMC showed that, while both provide extremely accurate results, PACE does the job in seconds, while MCMC takes hours. In criminal investigations, Adelman adds, those kinds of accuracy and time differences matter.
The key to PACE’s success is machine learning, which Adelman defines as having four parts:
- the model that makes predictions (in this case the number of contributors in a sample)
- features, or high-value information, in the vast sea of data (or, as Adelman says, “the signal amid the noise in the data set”)
- an algorithm that uses features from the data set to construct a model
- a training data set that the model uses to learn or to select those statistical and data-mining techniques best suited to solve the problem at hand.
“In other words,” Adelman says, “when it comes to utilizing the massive amount of available information in the DNA profile, machine learning does all of that computational heavy lifting up front during the creation of a model that is then passed on to the end user.”
Both Adelman and Marciano say they couldn’t have succeeded without a comprehensive training data set, which included major publicly available contributions from the Boston University School of Medicine as well as many laboratories and corporations. “That support was critical,” Marciano asserts.
Syracuse University has licensed the software to NicheVision, Inc., of Akron, Ohio, and along with Marciano and Adelman is currently commercializing the PACE software. As Adelman and Marciano continue to actively participate in the development of PACE, they have also received funding from the National Institute of Justice to look at a method for deconvoluting DNA mixtures, or separating out each individual’s DNA from a mixed sample. “The current gold standard within the community uses a Bayesian hierarchical model that relies upon MCMC,” Marciano explains. “While that has been implemented well, we believe that machine learning can speed things up here as well.”
Hair Apparent

Unlike the DNA in mixed samples, strands of hair can be easily separated from one another, and a new approach is now using hair for protein-based identification.
“With hair, we’re still looking at DNA, but we’re just looking at it in a radically different way, and that’s in terms of the signatures it leaves in proteins,” says Glendon Parker, Ph.D., who was a biochemist at the Lawrence Livermore National Laboratory (LLNL) Forensic Science Center (Figure 3). Research groups at LLNL and the startup company Protein-Based Identification Technologies of Orem, Utah, of which Parker is founder and CEO, are developing the approach, which centers on a class of DNA mutations that result in sequence changes within hair proteins (Figure 4). Known as nonsynonymous single nucleotide polymorphisms, or nonsynonymous SNPs, these mutations occur with different frequencies in individuals and therefore provide reasonable powers of discrimination for identification purposes [3]. “In a way, we’re doing DNA typing, but we’re doing it by looking at the fingerprints that DNA has in proteins, and then using those fingerprints to predict back to that DNA,” Parker explains.
Now with the University of California–Davis, Parker believes that the predictive powers of the technique depend on the variations in the DNA and the ability to extract that information from a sample. “At this point, using material from a single hair that Livermore received as a real-world sample from the FBI, we showed a power of discrimination of one in 90.” Parker explains that this is typically more than sufficient because such data are used not to identify one person from the general population but to consider a small pool of suspects.

To develop the methodology, the LLNL research group had to build on existing proteomics techniques to process hairs, which have very robust, highly cross-linked protein structures, and adapt standard proteomics tools to seek out nonsynonymous SNPs. “We were able to develop some additional reference databases that actually can detect those changes, and then we were able to incorporate them into our workflow,” Parker explains.
Currently, work is under way to increase sensitivity by expanding the number of mutations, or markers, the methodology seeks out. “Right now, we are at about 180 markers that have been validated, and it’s growing all the time,” he reports. “We’re also starting to develop more targeted approaches that use a particular type of mass spectrometry, called multiple reaction monitoring, or MRM, that is even more sensitive at detecting peptides in a sample and more quantitative.” Parker and the LLNL research group are also concentrating on validating and refining their statistical approaches and tools to the point of commercialization. “There are many validation steps that need to go into this, because if a law-enforcement agency is going into a courtroom, they want to make sure the evidence is of a very high quality to justify putting someone in jail.”
Parker conjectures that this approach might be useful outside of forensics, too. “Since we’re taking changes in peptides that we detect through proteomics and then going back to make a prediction about someone’s DNA, it’s feasible that you could set up large-scale processes to look for things like genetic variations that could be associated with genetic disease.” This runs contrary to how clinical biomedicine currently works,” he says. “The process now is to look in the DNA first, and, if you find something, you go on to look in the mass spectrometry or proteomic data, so it always goes from DNA to protein or peptide. What we’re doing is the opposite, and that’s actually pretty novel and quite interesting.”
Forensic Face Lift
Another innovative technology that is changing the capabilities of forensic investigators is one that can take a DNA sample and report the probability that a particular suspect has a certain eye, hair, and skin color and a particular facial shape, as well as his or her ancestry. Called Snapshot [4], this technology “can allow investigators to go back to their suspect lists and reprioritize them to spend their time focusing on the people who match these characteristics,” says Ellen McRae Greytak, Ph.D., director of bioinformatics for Parabon NanoLabs, Inc., of Reston, Virginia (Figure 5).
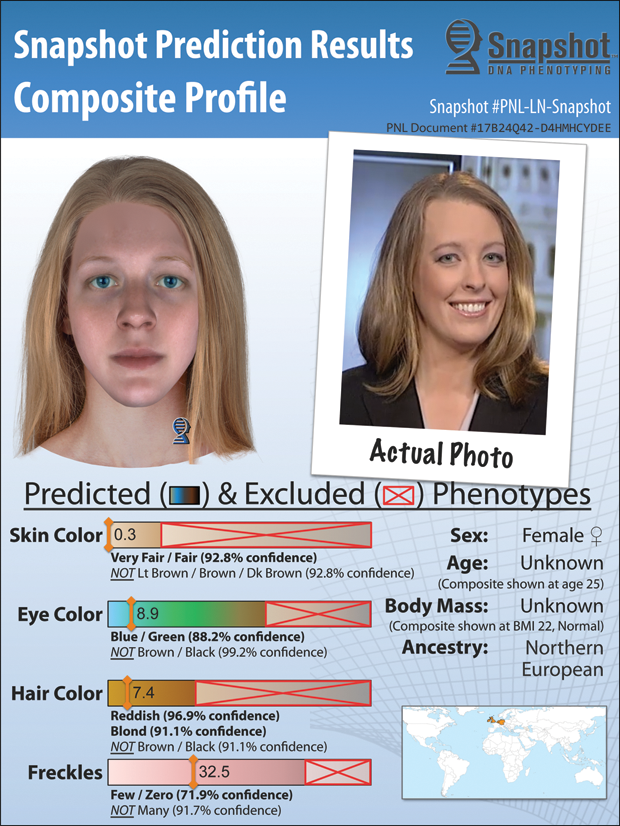
To illustrate its capabilities, Greytak describes how the company worked with police investigators to help with an unsolved double homicide that had occurred a few years earlier. “We did a Snapshot prediction based on a crime scene sample, and we were able to predict specifically that this was likely someone who had one Latino and one European parent. That was a light-bulb moment for the investigators, because they knew a person who fit that description,” she recalls. “After that, they took DNA from the suspect, it matched, and they were able to close that case very quickly.” Other cases are under way, although she can’t comment on them as they are still within the court system and therefore not yet public.
In building Snapshot, Parabon began by pulling together a database that includes both DNA and information about phenotype from thousands of subjects, so they could begin correlating DNA markers with certain physical traits. “For example, if there are a thousand people with blue eyes and a thousand people with brown eyes, we can look through about a million different markers in each person and find which are the ones that consistently differentiate these two traits,” Greytak explains. From there, deep data mining combines with advanced machine learning and an evolutionary search algorithm to construct predictive models that look for subsets of markers to establish the likelihood that a person will have a certain eye color. “Of course,” she adds, “it gets more complicated than that, because people have green or hazel or dark brown eyes, but that’s the idea.”
Interestingly, some physical characteristics cannot be calculated with Snapshot. “When we first started, we didn’t really know which traits were going to be strongly predictable. We thought things like handedness—left- versus right-handed—would be simple, but we found that we currently can’t predict either with high accuracy,” Greytak says. Height was also problematic. “Even though height is very strongly genetic, there are something like 100,000 different markers involved in height, and many of them may be rare enough that they don’t even show up in our data, so we’re not yet able to predict it.”
The Snapshot technology requires as little as a nanogram of DNA, which can be extracted from even a tiny swab of blood or other biological material. Samples go through a screening process to determine how much DNA is present, how old it is, how it’s been stored, and whether it is a mixed sample (most of the DNA from a mixed sample must be from the person of interest). The sample arrives at a clinical lab for microarray processing, and the resulting data go to Parabon, where Snapshot makes its predictions.
“Then, we very clearly brief the detectives about exactly how to use this information,” Greytak clarifies. “We present everything with a confidence value, and we try to focus on exclusions, so we might tell the investigators that we have 70% confidence that this person has blue eyes instead of green eyes, and 99% confidence that this person does not have brown eyes. That means investigators should start looking for people with blue eyes on their suspect list, then go to the green-eyed people, and finally, if they don’t find anyone there, look at the brown-eyed people.”
Besides narrowing down suspects, Snapshot has a range of other applications. While the technology can predict the shape of the face and apply skin, eye, and hair color, it cannot foresee those traits that change, such as hairstyle, age, and weight. At the same time, however, Snapshot’s predicted traits can be combined with other known information, such as the age of a cold case or general witness descriptions, so that a forensic artist can produce a composite drawing that advances the age correctly, adjusts the hair length or facial fullness, or includes other details.
Such composite drawings can be especially helpful for unidentified remains. “If a body is found with a skull,” Greytak says,” a forensic artist can do a black-and-white skull reconstruction, while we can independently do a Snapshot prediction to come up with the person’s pigmentation, eye color, and hair color to produce a full-color composite of that person.”
Beyond predicting appearance, Parabon is also applying its machine-learning approach to the problem of detecting distant kinship between pairs of subjects. Thus far, they are able to distinguish out to sixth-degree relatives (second cousins once removed) from unrelated pairs. “When unidentified remains are discovered, even if a possible identity is known, sometimes there aren’t appropriate living relatives available for traditional testing,” Greytak explains. “Our algorithms extend the distance over which relatedness can be detected from DNA, while maintaining an extremely low false-positive rate.” This technology is being developed in collaboration with the U.S. Armed Forces DNA Identification Laboratory, which is charged with identifying the remains of soldiers from past conflicts.
Faster, Better Forensics Tech
While some of these technologies may indeed lead to advances in other areas, researchers say the forensics field has many needs of its own. “One thing about forensics that I think is misunderstood sometimes is that the technologies we are using today are not the most novel or cutting-edge technologies. Instead, they are what works well enough and what we know like the back of our hands,” Marciano remarks. Field investigators do not necessarily gravitate toward the latest technologies, and “it can take a lot of effort to get laboratories to adopt new methods, even though they’re very sound and may even be much better.”
Palmbach completely agrees. “You’d be amazed how difficult it is to convince people to make the investment in technology and try something new. We can draw out the benefits for them, it can be so logical, but I think it’s going to take a lot of real success stories before people in the forensics community will understand and accept the real inherent value in these new technologies.” Once law enforcement is on board, however, change will come quickly, he contends. “All these things will be of tremendous value for traditional police work, and they will lead to increased solve rates; reduced costs because the police and judicial system are going to get scientifically verified information much more quickly; and reduced wrongful convictions.”
He adds, “The system will be better, faster, and more efficient.”
References
- Forensics Technology, M.S. University of New Haven. [Online].
- M. A. Marciano and J. D. Adelman, “PACE: Probabilistic Assessment for Contributor Estimation—A machine learning-based assessment of the number of contributors in DNA mixtures,” Forensic Sci. Int.-Gen., vol. 27, pp. 82–91, Mar. 2017.
- G. J. Parker, T. Leppert, D. S. Anex, J. K. Hilmer, N. Matsunami, L. Baird, J. Stevens, K. Parsawar, B. P. Durbin-Johnson, D. M. Rocke, C. Nelson, D. J. Fairbanks, A. S. Wilson, R. H. Rice, S. R. Woodward, B. Bothner, B. R. Hart, and M. Leppert. Demonstration of protein-based human identification using the hair shaft proteome. PLoS One. [Online]. 11(9).
- Parabon Nanolabs. Snapshot: DNA phenotyping, ancestry & kinship analysis. [Online].



