We live in an age of plentiful information, collected continuously by pervasive gadgetry, distributed through digital and social networks, and mined deeply by ever-more- powerful analytics systems. And yet, one of the things we know the least about is our bodies. When it comes to our own health, we are driving blindly. Modern medicine has clearly been remarkably successful, as evidenced by continually growing life expectancies. For example, the number of people 65 and older in the United States has seen a steady increase over the last century, rising from 3.1 million in 1900 to 41.4 million as of 2011—and is expected to grow to 80 million by 2040. Concurrently, the number (and fraction) of people suffering from chronic conditions is also growing, from 44.7% in 1995 to 47.7% in 2015—and expected to rise to over 49% by 2030 [1]. According to the U.S. Centers for Disease Control and Prevention, the vast majority of healthcare costs (86%) are spent on treating and managing chronic disease. Indeed, the medical field today focuses almost entirely on care after illness: modern medicine is largely reactive, waiting until the system fails before taking action. While the field of dentistry has fully embraced regular preventive evaluation and treatment with great success, most other medical fields have yet to do so. Yearly physical evaluations are recommended, but they rely on very limited data collection. As long as there are no symptoms, current standard medical practice largely foregoes testing (laboratory, imaging, etc.) as a screening method to detect disease before it manifests itself.
Changing a Perplexing Situation for the Better
This is a rather perplexing situation. It is as though we never did maintenance on our cars and just drove until they broke down, on the premise that one shouldn’t proactively test and fix problems before they reveal themselves. Would it make sense not to have an engine temperature readout and just let the engine overheat before we notice? Would it make sense not to check the brake pads until they fail—and let a car’s failure to stop when needed be the first symptom?
As a result of driving our bodies blindly, we simply don’t know when we may become chronically ill. But what if this were different? What would the practice of medicine (and its associated costs) look like if we routinely obtained detailed and multifaceted data about our bodies? Clearly, early detection of a disease should provide the best opportunity to reverse its course and prevent it from escalating and becoming chronic. As with car maintenance, this requires regular collection of data.
The Multiple Needs of Proactive Medicine
While regular data collection is crucial, it is not sufficient. Larry Smarr, one of the pioneers of the Quantified Self movement, has been collecting and manually curating a rich and growing set of data about his own body and health (see the article “The Quantified Patient Checks In” on page 4.). At one point, he noticed an unexplained spike in C-reactive protein (a blood marker of inflammation he had been following), which had reached values significantly outside normal range. When he presented these troubling findings to a physician, he was asked, “Do you have a symptom?,” then sent away without treatment, further investigation, or guidance on how to proceed, beyond “Come back when you have a real symptom.” Not long after, he indeed had an acute and potentially life-threatening event that required urgent care. This would have been prevented if the physician had treated the available information (a measured endophenotype) as an actionable indicator of health status.
To achieve proactive medicine, we thus need data, their integration into a powerful analytic system, and a significant change of current medical practice into a system that is personalized, predictive, preventive, and participatory: P4 medicine (also known, as pioneered by Leroy Hood, as a systematic approach.)
The Pioneer 100 Feasibility Study
Smarr’s pioneering work makes a compelling case for the utility of a data-driven medical system. What would happen if his method were applied at the population level? Given longitudinal and multifaceted information about the health status of many individuals, would it be possible to sustain their health, promote their wellness, and demystify disease? Would this lead to a democratization of health care [2]?
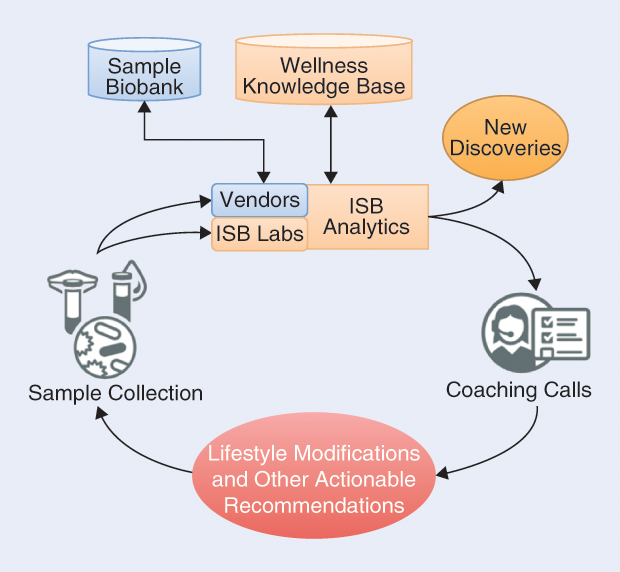
To evaluate this potential, the Hood-cofounded Institute for Systems Biology (ISB) in Seattle, Washington, initiated a pilot project to test the feasibility of collecting and interpreting multifaceted data for many individuals, providing them with actionable opportunities based on the data, and coaching them on ways to improve their health and wellness. Specifically, we recruited a group of 108 willing individuals (referred to as “pioneers”) into an institutional review board–approved study led by Prof. Hood and Prof. Nathan Price. The study design embodied a wellness cycle (schematically presented in Figure 1) with four stages:
- collection of biosamples from the individual
- experimental measurement and computational analysis
- communication of results and recommendations to the individual (via coaching calls)
- implementation of these recommendations by the individual.
These cycles repeat, leading to the accumulation of longitudinal data from which the individual’s wellness can be modeled and optimized.
The pioneers, who ranged in age from their 20s to over 88, were 58% male and 42% female and largely Caucasian. Their mean body mass index (BMI) was 25.2 kg/m². We launched the study in March 2014, and it lasted nine months. For each participant, we collected blood, urine, saliva, and stool samples at three-month intervals. From these samples, we obtained detailed information about their genetics by whole-genome sequencing (only in the first round of sampling, as the genome is not expected to change over time), on their endophenotypes (metabolites, proteins, and other molecules as observed in the blood and saliva), and on their gut microbiome. Participants also engaged in continual self-tracking and lifestyle monitoring via digital pedometers and questionnaires.
Our project management team, led by Sean Bell and Kristin Brogaard, Ph.D., encountered many organizational challenges relating to sample collection and processing, interfacing with analysis vendors, providing tools and service to the participants, and more. Successfully addressing these technical challenges provided insight into the mechanics of implementing a streamlined wellness service that adjusts itself to the personal needs of each individual and at the same time scales up to population level.
To help make sense of the resulting complex data, we organized the set of measurements into four health quadrants relating to diabetes risk, cardiovascular disease, inflammation, and nutrition. To provide the pioneers with the results of these analyses, regular phone conversations were scheduled with Sandi Kaplan, M.S., R.D., a wellness coach who helped them interpret the most important findings and discussed ways to act on the results and elicit positive change. Prior to these discussions, Craig Keebler, M.D. (who acted as study physician), reviewed all the data.
Personalized Data Clouds
We refer to the collection of data on each individual as each person’s “personalized, dense, and dynamic data cloud,” because it includes multifaceted, longitudinal information about his or her own health status. Clearly, in-depth analysis and interpretation of such data clouds pose challenges and offer many avenues for technology development in the future.
Even so, the initial review of the data collected for each pioneer yielded some surprises at the individual level. Based on the combination of their genetic profile and the clinical labs, the study physician determined that two pioneers had hemochromatosis—a potentially deadly disease easily treated by regular blood donations to reduce the iron stores. Left untreated, this disorder could lead to cartilage damage, liver cancer, diabetes, and heart disease. One participant already had cartilage damage from his undiagnosed disease; subsequent family genetic testing detected other family members at risk. By simple bloodletting, his ferritin levels were restored to normal range in fewer than three months. At the cohort level, we could observe the correlation between the number of copies of the genetic variants causing hemochromatosis and the ferritin levels in the blood of the pioneers.
The correlation between genetic predispositions and measurements also held for more complex phenotypes. Because we obtained whole genome sequence data for all study participants, we could directly observe the genotypes at all sites known to contribute to disease risk, without the need for imputation from partial data. For example, by integrating 59 previously identified genetic variants [3], each of which has a small effect on the risk of elevated low-density lipoprotein (LDL) cholesterol, we could compute for each pioneer his or her individualized, polygenic risk score. Across the cohort, we again saw an increase in levels of LDL cholesterol measured in the blood, as a function of increasing genetic risk.
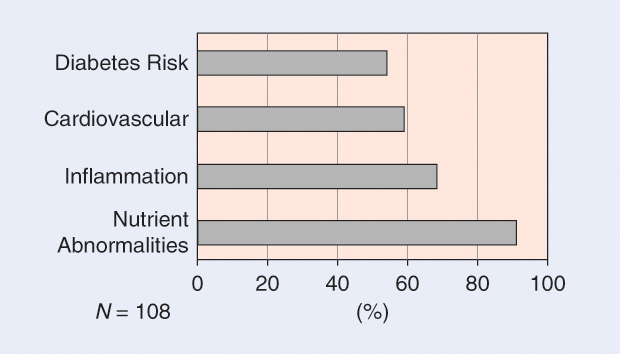
Cohort-level statistics on all measurements also revealed that this cohort of 108 largely healthy participants (some of whom had chronic diseases) had a high rate of initial lab results outside normal range. This ranged from 54% of pioneers having measurements placing them in the prediabetic to diabetic status to 91% of pioneers having at least one nutrient abnormality, mostly driven by vitamin D deficiency (Figure 2).
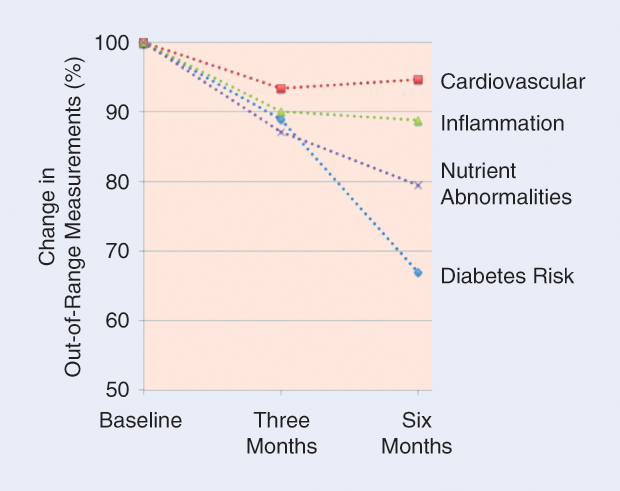
Based on these results, 100% of the pioneers received actionable recommendations via the coaching calls. By the end of the study, the frequency of out-of-range measurements had decreased in all four quadrants, with reductions relative to baseline of 33% for diabetes risk, 6% for cardiovascular disease, 12% for inflammation, and 21% for nutrition abnormalities (Figure 3).
New Discoveries
Lacking detailed information about each individual, it is customary to evaluate measured observations (e.g., of LDL cholesterol) relative to reference ranges, frequently stratified by age, sex, and perhaps the individual’s BMI. In our study, we collected significantly more detailed data for each individual, allowing our analytics team, led by Andrew Magis, Ph.D., and John Earls, to integrate heterogeneous data types into personalized and predictive models. We used ordinary least-squares regression to predict the measured levels of LDL cholesterol given various combinations of observations. We found that while the simple combination of age, sex, and BMI was not sufficient to predict the observed LDL cholesterol levels in the blood, the addition of genetic risk and observed gut microbiome taxa (in particular, Proteobacteria) yielded a statistically significant predictor. Its accuracy further improved when we excluded from analysis individuals taking cholesterol-lowering medications.
We similarly compared all measured analytes, computed polygenic risks for 130 genetic traits along with other data, and identified (after age and sex adjustment and stringent multiple-testing correction) 3,470 significant correlations across data types, connecting 766 different individual measurements (metabolites, clinical laboratory tests, genetic traits, microbiome taxa, and proteins). This yielded a large correlation network that we further studied using community analysis [4]. Several of the communities thus identified correspond to established knowledge, e.g., the involvement in cardiovascular health of analytes such as C-peptide, triglycerides, insulin, homeostatic model assessment of insulin resistance, fasting glucose, high-density lipoprotein cholesterol, and small LDL particle number. It is reassuring that such connections arise even from such a limited size cohort. Further analysis of this correlation network reveals many additional significant connections, some of which can be tentatively explained by considering the literature. Other findings may represent novel discoveries; it will be important to evaluate whether their statistical strength survives the expansion of the data set to include more individuals. Such analysis is currently under way.
The Future of Health Care
Even in this limited cohort of 108 individuals, our study uncovered diseases (hemochromatosis, diabetes), identified and removed abnormal levels of toxins (lead, mercury), and supported the management and reversal of chronic conditions (inflammation). During the course of this study, the pioneers were deeply engaged, providing data through questionnaires, contributing multiple samples for analysis, discussing the findings and the actionable recommendations with the wellness coach, participating in events, and, finally, offering feedback on the entire experience. Three recurring themes were apparent from this feedback. First, participants realized that the genome does not control their health destiny but rather frames the potential; second, that given the proper data, they can take control of their own health; and, finally, that people are not as well as they think: everyone had multiple actionable possibilities for enhancing his or her health.
The personalized, dense, and dynamic data clouds are not just an accumulation of data. For each participant, the integration of multiple data types enables improved interpretation of the findings. Our participant engagement team, led by Jennifer Lovejoy, Ph.D., also found that the combined evidence from more than one data type (e.g., genetic propensity and abnormal clinical findings) was more effective at eliciting behavior change than individual out-of-range findings. Further, the combination of data clouds from many individuals enabled both validation of previous biomedical knowledge and observation of novel statistical connections to be formally tested. The personalized, dense, and dynamic data clouds are thus a technology platform that will enable new discovery as well as new approaches to stratification of patients—a requisite for precision medicine.
The personalized data clouds and their integration into a learning engine for systems medicine represent the foundations of the personalized, predictive, preventive, and participatory medicine of the future. By measuring a broad range of parameters longitudinally, it will be possible to recognize the earliest signs of disease [5]. If the course of disease progression can be reversed at its earliest and most malleable point, healthcare costs will be significantly reduced. This will lead to major changes in the pharmaceutical, diagnostic, biotechnology, and nutrition industries and usher in an era in which wellness care democratizes access to long and healthy lives.
References
- S. Mattke, L. Klautzer, T. Mengistu, J. Garnett, J. Hu, and H. Wu. (2010). Health and well-being in the home. RAND Corporation. Santa Monica, CA. [Online].
- L. Hood and N. D. Price, “Demystifying disease, democratizing health care.” Sci. Transl. Med., vol. 6, no. 225, p. 225ed5, 2014.
- Global Lipids Genetics Consortium, “Discovery and refinement of loci associated with lipid levels,” Nat. Genet., vol. 45, no. 11, pp. 1274–1283, 2013.
- M. Girvan and M. E. J. Newman, “Community structure in social and biological networks,” Proc. Natl. Acad. Sci. U.S.A., vol. 99, no. 12, pp. 7821–7826, 2002.
- R. Chen, G. I. Mias, J. Li-Pook-Than, L. Jiang, H. Y. Lam, R. Chen, E. Miriami, K. J. Karczewski, M. Hariharan, F. E. Dewey, Y. Cheng, M. J. Clark, H. Im, L. Habegger, S. Balasubramanian, M. O’Huallachain, J. T. Dudley, S. Hillenmeyer, R. Haraksingh, D. Sharon, G. Euskirchen, P. Lacroute, K. Bettinger, A. P. Boyle, M. Kasowski, F. Grubert, S. Seki, M. Garcia, M. Whirl-Carrillo, M. Gallardo, M. A. Blasco, P. L. Greenberg, P. Snyder, T. E. Klein, R. B. Altman, A. J. Butte, E. A. Ashley, M. Gerstein, K. C. Nadeau, H. Tang, and M. Snyder, “Personal omics profiling reveals dynamic molecular and medical phenotypes,” Cell, vol. 148, no. 6, pp. 1293–1307, 2012.



