2019 was a revolutionary year in the field of chemistry as the Royal Swedish Academy of Sciences awarded the Nobel Prize in Chemistry 2018 to three brilliant chemists (Figure 1): Dr. Frances H. Arnold—“for the directed evolution of enzymes”; Dr. George P. Smith and Sir Gregory P. Winter (jointly)—“for the phage display of peptides and antibodies” [1]. Their phenomenal contribution in controlling the evolution of enzymes and binding proteins and using them for the benefit of humankind has revolutionized the industry and academia alike for the past few decades.
The laureates

Dr. Frances Hamilton Arnold is currently the Linus Pauling Professor of Chemical Engineering, Bioengineering and Biochemistry at the California Institute of Technology, Pasadena, CA, USA. She received the B.S. degree in mechanical and aerospace engineering from Princeton University in 1979, where her focus of research was solar energy. After her graduation, she worked as an engineer in different countries and at the Solar Energy Research Institute in Colorado before joining the University of California Berkeley, CA, USA, for a Ph.D. in chemical engineering. After completing her post-doctoral research in 1986, she joined the California Institute of Technology as an assistant professor.
During her career as a professor, she has pioneered “directed evolution of enzymes,” which is harnessing the principles of evolution for engineering proteins by gene mutation and artificial selection, to create improved biocatalysts that can solve several major global issues in agriculture, sustainable energy, and health care. She is also the co- inventor of 60 patents and co-founder of Gevo, Inc., a company making renewable chemicals and advanced biofuels. Apart from the prestigious Nobel Prize, Dr. Arnold is the recipient of numerous awards, including the Sackler Prize in Convergence Research (2017), Millennium Technology Prize (2016), the National Medal of Technology and Innovation (2013), and the NAE’s Charles Stark Draper Prize (2011), among many others. She has been elected to prestigious memberships in all three U.S. National Academies of science, medicine, and engineering [2].
Dr. George Pearson Smith is currently the curators’ Distinguished Professor Emeritus of Biological Sciences at the University of Missouri, Columbia, MO, USA. He received the bachelor’s degree from Haverford College, Haverford, PA, USA. After a stint as a teacher, he received a doctorate in bacteriology and immunology in 1970 from Harvard University. Following post-doctoral research at the University of Wisconsin, Madison, WI, USA, he joined the University of Missouri as faculty in 1975. During his sabbatical at Duke University from 1983 to 1984, he came up with the idea of using a simple process called “phage display” to identify an unknown gene for a known protein, for which he was awarded the Nobel Prize in Chemistry in 2018.
Using Smith’s concepts of phage display, another scientist, Sir Gregory Paul Winter, developed directed evolution of antibodies, which can bind to a specific protein involved, for example, with a human disease. Sir Gregory Paul Winter graduated from Trinity College, Cambridge, U.K., in 1973. He received the Ph.D. degree in 1977 from the MRC Laboratory of Molecular Biology, Cambridge, for research on the amino acid sequence of tryptophanyl tRNA synthetase from the bacterium Bacillus stearothermophilus. Winter completed his post- doctoral work at Imperial College London, London, U.K., and the Institute of Genetics in the University of Cambridge. A large part of his research career has been based entirely at the MRC Laboratory of Molecular Biology and the MRC Center for Protein Engineering, Cambridge. His research on protein and genetic engineering has led to successful spin-off companies including Cambridge Antibody Technology (acquired by AstraZeneca), Domantis (acquired by GlaxoSmithKline), and Bicycle Therapeutics [3].
Directed evolution of enzymes: Rewriting the code of life
Over billions of years, our planet Earth has been able to sustain numerous living organisms due to a process called evolution. Evolution has led to organisms being able to survive harsh environments by mutation of their genetic code and elimination of their weaker progeny by natural selection. Arnold adapted this concept of natural evolution and brilliantly replicated the procedure in her laboratory for artificially controlling and speeding up the evolution process in enzymes. These “fantastic-biocatalysts” offer an environment-friendly alternative to harmful chemical catalysts and have been practically implemented in various industries over the past two decades.
Enzymes are complex molecules consisting of thousands of combinations of amino acids. Arnold’s initial approach toward rebuilding enzymes with new properties turned out to be extremely difficult because of their vast number. Instead, Arnold resorted to the smart idea of creating a random mutation in the genetic code of a specific enzyme and introducing these mutated genes to bacteria that produced different varieties of this enzyme with mutations. These different varieties were then screened for a desired property. Those enzymes that are more efficient in catalyzing a desired chemical reaction are chosen (similar to the principle of natural selection) and subjected to new random mutations. After several iterations, the desired efficiency is achieved. The entire workflow of directed evolution of enzymes is demonstrated in Figure 2.
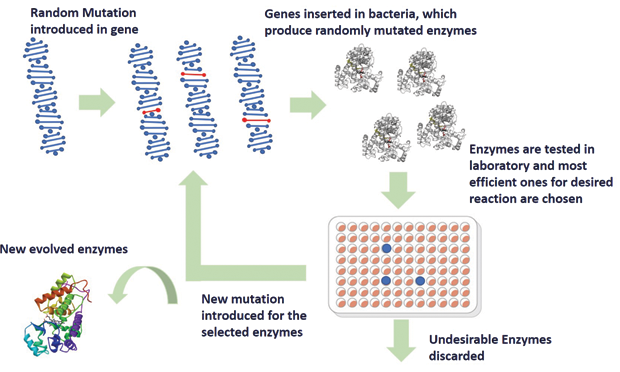
In the seminal work published in 1993 [4], Arnold demonstrated the directed evolution of an enzyme called subtilisin E. The evolved subtilisin E showed 256-fold higher activity in its unnatural environment, i.e., in an organic solvent, compared to the wild type. This was a benchmark experiment, which initiated a new era of directed evolution of enzymes that ultimately led to the creation of several useful biological systems like enzymes with new properties, new metabolic pathways, renewable biofuels, and environment-friendly chemical synthesis. Directed evolution, in addition to improving the function of existing proteins, enabled the creation of new enzymes with protein functions for which no natural enzyme existed. For example, the enzyme Cytochrome P450, whose natural function is metabolism, could be modified to perform an entirely new function—to naturally insert oxygen atoms in the drugs we ingest [5]. This enables designing ecofriendly drugs and pesticides. Evolved enzymes that transform sugar into isobutanol, an organic compound that can be utilized for environment-friendly alternative to fuel and plastic, has also been demonstrated. The technique of directed evolution has successfully brought new chemistry into the biological world with the promise of a greener world.
Manipulating phages for tailoring antibodies
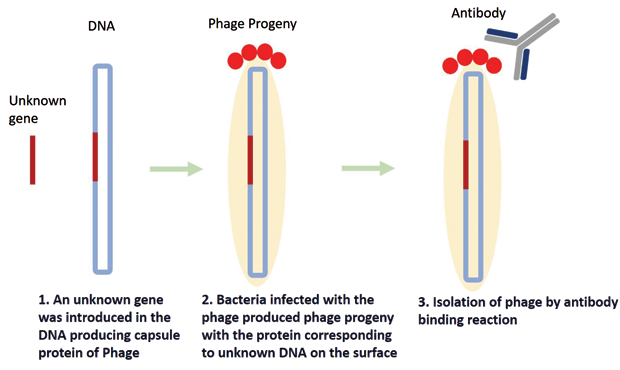
Phages are viruses consisting of a DNA or RNA genome encapsulated by proteins. Phages attack bacteria by injecting their genetic material, which then takes over the bacteria’s metabolism. The phage’s genetic material replicates, and the encapsulating protein accumulates inside the bacteria. The bacteria then extrude several copies of the progeny phage. In 1985, Smith used this simple mechanism of phages to find an unknown gene for a known protein [6]. He used a piece of unknown gene and combined it with the gene for one of the proteins in the phage capsule of a filamentous phage. When the progeny phages were produced, the protein corresponding to the unknown gene appeared on the surface of the phage along with the capsule protein. This process is known as “phage display” (Figure 3). At the next step, by using a suitable antibody, the phage carrying the targeted protein could be fished out. Thus, phages acted as a link between genes and proteins, and unknown genes responsible for the expression of a specific protein could be easily identified.
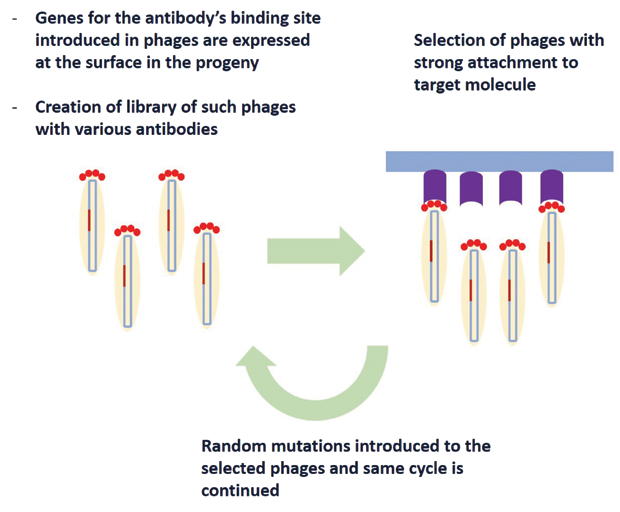
A direct consequence of phage display is engineering antibodies. Initially, researchers tried to extract antibodies for a specific disease by injecting mice with a target protein for that specific disease. Several limitations of such a method led Winter to start investigating the potential of Smith’s phage display for engineering antibodies, where the gene responsible for the antibody’s binding site was combined with the genetic material of phage’s capsule protein. This led to the expression of the antibody’s binding site on the surface of the phage. A library of phages with various types of antibodies on their surface was created, and antibodies that attach to the target molecule are then fished out from this library. These are then subjected to random mutations, creating a second generation of phages with antibodies on the surface that have stronger attachment to the target molecule. After a few iterations, the antibodies generated had an extremely high level of specificity to the target molecule, a method known as “directed evolution of antibodies.” Figure 4 demonstrates this basic concept schematically. Winter’s demonstration of phage display of antibodies [7] in 1990 revolutionized the pharmaceutical industry and led to tailoring of protein, peptides, and antibodies with endless possibilities.
How directed evolution of enzymes and antibodies ended up in laundry detergent and beyond
The discoveries that led to the Nobel Prize in Chemistry 2018 have revolutionized the fields of pharmacy, energy, and chemical synthesis. Over the past few decades, Arnold’s directed evolution of enzymes and Smith and Winter’s phage display of antibodies have demonstrated unprecedented possibilities for a greener and disease-free planet.
Arnold’s directed evolution of enzymes has been adopted widely in designing drugs more efficiently by the pharmaceutical industry. In addition, being able to tailor enzymes has led to the development of biofuels— a greener alternative to non-renewable sources of energy. Directed evolution has also enabled replacing traditional chemistry, which often involves the use of toxic chemicals, with an environment-friendly alternative for the production of fertilizers, industrial solvents, detergents, etc. The evolved enzymes, which have higher level of specificity, higher tolerance to diverse environments and limited redundant reactions, have enabled the discovery of new reactions that are unknown to the biological world. Realizing the importance of this method, in 2005 Arnold co-founded Gevo, Inc., which focuses on manufacturing renewable chemicals and biofuels. In the past two decades, directed evolution has shown its true potential, and it remains to be seen what other exciting possibilities it brings forward in the future.
While directed evolution of enzymes has made its mark in the chemical and energy industry, phage display of peptides and antibodies has revolutionized the pharmaceutical industry. It led to engineering antibodies for various diseases, including cancer. In order to use the potential of phage display for practical applications, Winter founded an antibody engineering company called Cambridge Antibody Technology. Winter’s research led to the discovery of a drug based on a human antibody called HUMIRA (adalimumab) that can halt inflammation in the body. HUMIRA was approved in 2002 to fight rheumatoid arthritis and is the world’s first fully human antibody [8]. The success of phage display of antibodies in developing the first human antibody triggered the pharmaceutical industry to use the technique for developing cancer antibodies. A cure for metastatic cancer has also been demonstrated. Antibodies to curing anthrax and autoimmune disease such as lupus have been approved. Many others are currently in clinical trials. The concept of phage display of antibodies holds promise for several other breakthroughs in the health care industry.
The discoveries of Arnold, Smith, and Winter mark a new era in tailoring proteins for the benefit of mankind. The last few decades have shown time and again, through various applications, the enormous technological impact of the directed evolution of enzymes and phage display of antibodies, but is merely the tip of an iceberg.
References
- “The Nobel Prize in Physics 2018.” [Online].
- F. Arnold and K. A. Macuare, “The NAI fellow profile: An interview with Dr. Frances Arnold,” Technol. Innov., vol. 18, pp. 79–82, 2016.
- Webpage of MRC Laboratory of Molecular Biology, Cambridge, U.K. [Online].
- K. Chen and F. H. Arnold, “Tuning the activity of an enzyme for unusual environments: Sequential random mutagenesis of subtilisin E for catalysis in dimethylformamide,” Proc. Nat. Acad. Sci., vol. 90, no. 12, pp. 5618–5622, 1993.
- P. S. Coelho et al., “Olefin cyclopropanation via carbene transfer catalyzed by engineered cytochrome P450 enzymes,” Science, vol. 339, no. 6117, pp. 307–310, 2013.
- G. P. Smith, “Filamentous fusion phage: Novel expression vectors that display cloned antigens on the virion surface,” Science, vol. 228, no. 4705, pp. 1315– 1317, 1985.
- J. McCafferty et al., “Phage antibodies: Filamentous phage displaying antibody variable domains,” Nature, vol. 348, no. 6301, pp. 552–554, 1990.
- S. Lawrence, “Billion dollar babies—Biotech drugs as blockbusters,” Nature Biotechnol., vol. 25, pp. 380–382, 2007.