A technology company in Gaithersburg, MD, USA, has developed a three-pronged approach to help researchers analyze microbiome samples. It includes a thorough microbiome database, an algorithm to search it, and a cloud platform to return useful information.
The database was the most time-consuming of the three parts, said Nur Hasan, Ph.D., molecular microbiologist and chief executive officer of EZBiome (Figure 1). Company researchers started off by conducting a study of the National Center for Biotechnology Information’s (NCBI) widely used public database of known bacteria, viruses, and other organisms in the gut microbiome. “We found up to 30% of the NCBI database entries are either misidentified or incompletely identified,” he said. So, the company’s researchers began what became a decade-long effort to build their own database [1], which he described as including 22,000 species that are “extensively curated and taxonomically validated.” Microbiome database development is a laborious undertaking, but since it is at the heart of microbiome analysis, he said, it was worth the effort.

Figure 1. Nur Hasan, Ph.D., molecular microbiologist and chief executive officer of EZBiome. The company has developed a suite of tools (www.EzbioCloud.net) to help researchers analyze microbiome tools. (Photo courtesy of EZBiome.)
With the database in place, they concentrated on an algorithm to serve essentially as a search engine that can match species in a sample to those in the database. They then went one step further by creating a cloud-based platform that employs precision taxonomy, which was initially developed in part by the founder of parent company CJ Bioscience, Seoul, South Korea [2], and is able to not only find exact species matches, but also identify and report taxonomic units that are not in the database. “Once that happens, we try to define it and add it to the database, and by doing so, we can classify more organisms very accurately.” The proprietary platform, available at EzBioCloud.net, is arranged with point-and-click functionality to make it as easy to use as possible, he noted.
“Apart from that, we developed unique gut-microbiome testing that takes the analysis to the next level,” said Hasan, noting that the company has collected data from more than 120,000 people who are either healthy or who have one of more than two dozen diseases (Figure 2). By applying machine-learning and artificial intelligence to that data, the company worked with 14 medical universities hospitals to generate a global microbiome health index that provides scores indicating the health of individuals’ microbiomes. “We validated it using two years of a citizen science project that included 10,000 individuals,” he said.
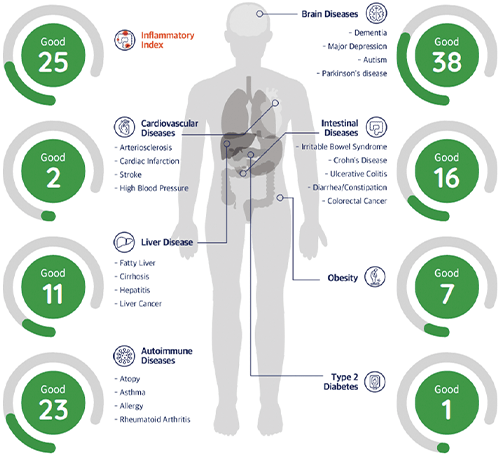
Figure 2. EZBiome has also created a gut-microbiome test, currently for research use only, that provides insight into the state of a person’s microbiome. It is designed to not only identify an unbalanced microbiome, but also to predict associated disease risks (data from a test shown). EZBiome is working with a few companies to potentially commercialize the test. (Image courtesy of EZBiome.)
The test is not diagnostic but for research use only, he pointed out. Hasan envisions it being used as a way to spot an unbalanced microbiome and predict potential disease risks, as well as a means to promote lifestyle modifications to rebalance the microbiome to lower risks. EZBiome is working with a few companies to potentially commercialize the risk test.
As this work continues, company researchers have begun developing therapeutics for a number of microbiome-related diseases, as well as companion diagnostics for immuno-oncology. They are also collaborating with clinicians, physicians, researchers, and industries around the world to generate what Hasan calls a global microbiome database that will collect data from the diversity of people living in different areas, with various ethnicities or cultures, and who have different diets [3]. For the latter, he remarked, “The more samples we have, the more enlightened we will be, the more we’ll understand about the microbiome, and the faster we can discovery novel therapeutic solutions to revolutionize the medicine.”
References
- S.-H. Yoon et al., “Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies,” Int. J. Syst. Evol. Microbiol., vol. 67, no. 5, pp. 1613–1617, May 2017.
- J. Chun et al., “Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes,” Int. J. Syst. Evol. Microbiol., vol. 68, no. 1, pp. 461–466, Jan. 2018.
- H.-S. Oh et al., “Proposal of a health gut microbiome index based on a meta-analysis of Korean and global population datasets,” J. Microbiol., vol. 60, no. 5, pp. 533–549, May 2022.



