The human body is a microbe’s playground: interspersed among the 37 trillion cells of the human body [1] are at least ten times as many bacteria, fungi, viruses, and archaea. That amounts to 100 times as many microbial genes as human genes.
Although the benefits of microbes were not unknown to science, the human microbiome’s importance, and in some cases its necessity to human health, is a notion that is taking center stage and generating a surfeit of questions. In the past few years, the study of the microbiome has mushroomed as researchers excitedly explore what makes up these huge populations of microbes in and on our bodies, exactly what they are doing there, and how they can be manipulated to promote health and combat disease.
Hundreds of studies and many different approaches (see “Zeroing in on Microbes”) are now under way to unravel the relationships between the microbiome and diseases ranging from autism to cancer. “These are the early days in our understanding of the microbiome, but already you’d be blown away by how much we’re learning,” says Owen White, associate director of the Institute for Genome Sciences at the University of Maryland School of Medicine.
[accordion title=”Zeroing in on Microbes”]

The microbiome is a tough nut to crack. Part of the reason for this is that clinical samples contain all kinds of things, such as human cells, salts, and other materials, that aren’t part of the microbiome, and researchers haven’t always had a good way to separate the microbes from the nonmicrobes or to quickly and easily zero in on one microbe. Several researchers, however, are developing methods that will allow for those capabilities and, in so doing, hope to have an impact in the clinic and perhaps on the broader understanding of the spread of disease.
One researcher is Joseph Petrosino, Ph.D., director of the Alkek Center for Metagenomics and Microbiome Research at the Baylor College of Medicine, and founder of Metanome, Inc., both in Houston, Texas. “We are able to purify live organisms from clinical samples for further study,” Petrosino explains. To do it, he and his research team are working with engineers Lorenzo D’Amico, Ph.D., and Peter Gascoyne, Ph.D., at Advanced Electrofluidic Systems, LLC, to develop a system that combines, refines, and expands some existing technologies in such a way that they can pull out just the bacteria from clinical samples. “Basically, we remove contaminating particulates and ions that would interfere with downstream processing by using what we call membraneless dialysis along with sedimentation in an enclosed microfluidic system. We then couple that with dielectrophoresis, which exploits the intrinsic electrical properties resulting from the structural form and composition of cells to isolate them without the need for bioengineered tags or labels. We can subsequently recover the collected organisms just by turning off the current.”
With the collection of isolated bacteria, researchers can then apply other techniques, including single-cell analysis, which is an up-and-coming area that would allow sequencing of individual microbes. For his own work, Petrosino is especially interested in metabolic screens. “The cool thing that we are initially doing is introducing metabolites into the closed system, letting them incubate with isolated communities of organisms, and then retrieving the metabolites after that incubation time,” he says. “We can begin to ask what this community is doing at the metabolic level without having to go through a series of laboratory manipulations to do so.”
Although their platform is still being refined, they are starting to use the dielectrophoresis technique in collaboration with interested researchers and, at the same time, are bringing the system up to scale so they can use it on a greater number of samples simultaneously and on larger sample volumes. They are also pursuing commercialization for clinical applications, including identifying disease-causing bacteria in a time span of hours rather than the days currently required, Petrosino says. “This would speed the ability to diagnose infections and perhaps to even identify antibiotic-resistance mechanisms that may be present.”
While these avenues continue, Petrosino also hopes to expand the technology so that it can isolate viruses. “The ability to isolate viruses from host material for sequencing applications is a significant hurdle in the field, and, if we can do so using dielectrophoresis, that would be a huge advancement.”
Single-cell analysis also intrigues Paul Blainey, who believes it has much to offer microbiome studies [1]. Blainey, Ph.D., is an assistant professor of the Massachusetts Institute of Technology (MIT) Department of Biological Engineering and a core member at the Broad Institute of MIT and Harvard University. One reason he sees single-cell analysis as a good option for microbiome studies is that it doesn’t involve growing samples in a culture, which can be very time-consuming and can miss some difficult-to-grow microbes.
While metagenomics is also culture-independent, it involves extracting all DNA from a sample and analyzing it as a pool, Blainey explains. “What single-cell analysis adds is the ability to organize the DNA sequences—essentially genes—that you get back according to which organism type they came from,” he says. “For instance, we might want to just have a list of all the genes that are present in one particular strain of bacteria so we can see how they work together in functional pathways, such as how the bacterium metabolizes a substrate or expresses a certain level of virulence.”
Single-cell analysis also allows for the study of individual bacterial cells. “The different individual cells of a single type of organism are themselves not all identical, so to analyze those differences comprehensively, you have to go to the single-cell level,” Blainey says. The advantage becomes clear in antibiotic resistance. “If we look at the DNA sequence from individual cells, we can resolve the microbial phylogenies with an incredible degree of resolution in order to track how resistant microbes are spreading (whether they’re spreading) through an individual patient’s body, through a ward in a hospital, or through a city or larger geographic region,” he adds.
Blainey has been working on single-cell analysis since 2007, when he was a doctoral student in the Stanford University Bioengineering Laboratory of Stephen Quake, Ph.D. There, Blainey was part of a research group that built an integrated microfluidic lab-on-a-chip system that could select individual microbes using an optical tweezer (a focused beam of light that physically sorted microbes) and then perform whole-genome amplification reactions on the individuals of interest.
Blainey has expanded on that work at the Broad Institute, where he leads an effort to set up a streamlined workflow for getting single cells ready for sequencing. “The sample preparation is really where the bottleneck is now,” he says. The new workflow is a standard wellplate process designed for high throughput and low cost, and has just begun accepting samples to run.
The next steps are to optimize the amplification chemistries to improve genome recovery and reduce potential errors, such as reading chimeric sequences, and to refine an automated microfluidic device they’re developing for single cell and conventional sample preparation applications. “Currently, it’s a lot of manual work to go from a colony on a plate to a genome sequence, but we’ve demonstrated that this new technology can go straight from whole cells to a sequencing library, so we think this could enable some large-scale, isolate-sequencing projects that would benefit microbiome research and clinical microbiology,” says Blainey.
The clinical applications of microbiome analysis technologies are coming, according to Blainey. Just as genetic screening is beginning to help cancer patients obtain personalized therapy, sequencing technologies can also be applied in the infectious disease area to identify specific disease-causing microbes. Another benefit lies in epidemiology, he says. “It’s a tragedy that most of the microorganisms that are isolated from patients through culture-based diagnostics are just thrown away.” Blainey hopes that his streamlining technology will one day make it possible to sequence those strains, especially those that are antibiotic resistant, which will enable the tracking of resistance “around the country, around the city, around the hospital ward, and even from one body site in a given patient to another body site.”
Blainey adds, “Microbiome research and technology present a big opportunity for impact at the public health level.”
Reference
- P. C. Blainey, “The future is now: Single-cell genomics of bacteria and archaea,” FEMS Microbiol. Rev., vol. 37, no. 3, pp. 407–427. May 2013.
[/accordion]
The possibilities arising from the view of the microbiome as working with human cells and physiological pathways, and potentially fighting disease, have triggered a growing demand for research and technologies that can open windows to understanding. For example, using techniques such as high-throughput sequencing, scientists and medical experts are learning more every day). As knowledge grows, this new understanding may just help us live longer and better lives.
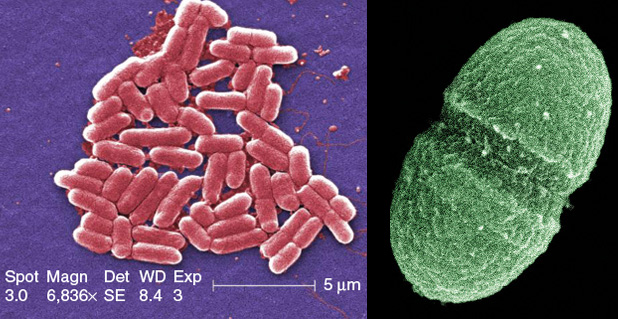
Right: Interspersed among the 37 trillion cells of the human body are at least ten times as many microbes. Many are harmless and potentially helpful, such as the bacterium Enterococcus faecalis (shown here), which resides in the human gut. (Photo courtesy of the U.S. Department of Agriculture.)
More Than Germs
Anton von Leeuwenhoek first brought attention to microbes back in the 1600s when he carefully made lenses for a microscope that provided views of an uncharted world. Using this revolutionary piece of equipment to sample his mouth and other parts of his body, he saw tiny organisms living everywhere he looked.
With his discovery came subsequent studies showing the connection between microbes and illnesses. “Since Leeuwenhoek first ground down those lenses and found bacteria, our perception of microbes has been primarily as pathogens: agents of infection and disease. As a result, most of our attention in research and medicine has been on microbes as germs,” says Lita Proctor, Ph.D., coordinator for the U.S. National Institutes of Health (NIH) Human Microbiome Project (HMP).

That prevailing belief really only began to change about a decade ago, she asserts. In 2003, the international research effort known as the Human Genome Project finished mapping the entire genetic blueprint of the human body, thanks to the maturing of new technologies to sequence DNA. “So of course, the larger scientific community was watching this incredibly moon-shot project to sequence the human genome. At the same time, microbiologists could see that the job wasn’t finished yet, so they contacted the NIH and said, ‘You really need to complete the other genetic component that we have, which is the collection of endogenous microorganisms that live in and on our bodies,’” Proctor recalls.
The microbiologists’ plea was buoyed by the availability of sequencing technology generated by the Human Genome Project. The technology was critical for microbiome studies because the vast majority of microbes aren’t accessible through the standard culture-based laboratory methodologies, where a technician might smear a sample of a microbe on a nutritional medium, watch to see whether the microbe survives and grows, and, if so, test the resulting colony’s response to other additions to the culture, such as potential antimicrobial agents. “If you look at the human microbiome, it turns out that about 70% are not culturable in that fashion, so we really did need sequencing technology to get at these communities,” Proctor says.
Added to the accessibility of sequencing technology, the price to do it was also dropping, adds White. “When we first sequenced the human genome, it cost billions of dollars to acquire the sequence of one person, and many different laboratories had to contribute. Now, we can generate the sequence of a human in an afternoon for US$2,000, so, all of a sudden, we’re presented with this new, incredibly powerful, and inexpensive microscope, if you will, that we can point in any direction and make a discovery.”
This confluence of factors prompted the NIH, through its umbrella organization The Common Fund, to establish the HMP in 2008. Its mission was to characterize the microbes that live in and on a healthy human body. Through this project, the perception that all microbes were bad-for-you germs was dispelled once and for all.
Making Use of the Microbiome
The HMP’s initial five years were focused mainly on determining which microbes comprise a healthy person’s microbial contingent, and how that contingent varies in different parts of the body, such as the gut microbiome versus the skin microbiome. Through the sequencing and analysis of samples taken from 242 healthy individuals, the many researchers and institutions participating in this first phase of the HMP provided a baseline description of the constituents of a normal, healthy microbiome, which was published in 2012 [2].
“There was this explosion of knowledge using sequence-based analyses that showed us the really rich, complex, and diverse community that lives all over the human body, and I do mean all over, because complex communities of bacteria, viruses, fungi, protozoa, and other kinds of microscopic life live in our guts, in our mouths, in our vaginal tracts (in women), on our skin, in our noses, in our lungs, and everywhere you look on our epithelial surfaces,” Proctor explains.
Beyond simply identifying which microbes live where, the project has expanded to its second phase, which includes illuminating the roles of microbes in disease. An important aspect is identifying which microbes are correlated with a particular illness or risk of illness and how drugs or other treatments affect microbes’ roles in diseases.
Matching up microbes and diseases, however, is a messy business. Not only are researchers furiously generating massive amounts of data on their own microbes of interest, but the confounding interconnectedness of physiological processes that is the norm in the human body also means that every researcher’s findings have potential implications for everybody else’s findings.
Ecologists are familiar with the dilemma. It’s impossible to truly understand an organism’s part in an ecosystem without considering everything that it affects and everything that affects it, and that can quickly turn a seemingly simple study into one that is overwhelming. “It should not be underestimated how the advances and the hurdles experienced by people in the ecology field over the years have impacted the growth in the microbiome arena,” says Joseph Petrosino, Ph.D., director of the Alkek Center for Metagenomics and Microbiome Research at the Baylor College of Medicine in Houston, Texas. “As we generate more data, and as the data sets become larger and more complex, the hope is that all of us in microbiology, ecology, mathematical modeling, and other fields will benefit by the development of new computational tools to deal with these big-data problems.”
Dealing with just these types of big-data issues is the job of the HMP’s Data Analysis and Coordination Center, according to White, who is the center’s principal investigator. “It’s fair to say that microbiome research is rivaling other disciplines, such as astronomy, physics, and meteorology, in terms of the amount of data that we deal with,” he says. “To give you an idea, when you generate a sequence from a microbiome sample and put it on a computer disk, just to open that file, uncompress the file, count all the nucleotides that are in the DNA sequence, and recompress it takes 30 minutes. In the last study that we did, we were dealing with more than 750 sequence files, so we had to deploy that information out on the grid just to do something as simple as count all the nucleotides.”
New tactics are helping to address these types of big data. In some cases, large computing grids and parallel systems are part of the solution. “That involves systems engineers who understand the relationship between the architecture of a central processing unit and file system. We’re also looking at using graphical processing units and different types of architectures, such as new Intel chips, to develop different algorithms and novel processing approaches,” White says. In addition, the Data Analysis and Coordination Center is helping researchers directly by offering tutorials that provide step-by-step instructions for performing data analyses, including the ability to perform automated sequence analysis from the user’s desktop [3]. The center is also providing access to all kinds of data, including the results from the HMP’s 15 demonstration projects [4] in which researchers are studying specific microbial populations, especially those related to human illnesses.
Another Twist
One more fly in the ointment for microbial studies is that the microbiome is exceptionally malleable, explains Chris Huttenhower, associate professor in the Biostatistics Department at the Harvard School of Public Health. Through the Broad Institute in Boston, he and researcher Ramnik Xavier of Massachusetts General Hospital are undertaking their own project to show just how much and how quickly it can change.

“A lot of early work in microbial communities was to treat it like a genome: you sequence and sequence, and you get some finished project, such as a genome assembly,” Huttenhower says. But that’s not how it works. “There is no ‘done’ when you’re sequencing the microbiome, because if you come back and repeat it tomorrow, next week, or next month, you’re going to get a different answer.” The reason is that a person’s complement of microbes changes on a day-to-day basis. Simple dietary differences will shift the bacteria in the gut because some will like the new nutrients and thrive, while others won’t and will die off.
Huttenhower and Xavier have specifically designed their study to track these changes and their effects on inflammatory bowel disease. “We are recruiting individual patients and will be following each patient over a year, so we’ll understand how the microbe evolves as their disease develops and how it responds to things like changes in treatments or in disease activity, Huttenhower says. The idea is to 1) understand at a basic, mechanistic level how the gut microbiome is perturbed in inflammatory bowel disease, 2) use that information to identify genetic points in the microbiome that can predict the response to treatments or the progression of the disease in individual patients, and 3) determine how to modify these genetic points to use them as therapeutic interventions.
As part of the HMP mandate, this project and others that are part of the HMP’s second phase have to produce more than just sequencing data, Huttenhower says. “On the one hand, that adds some complexity because it means that we have to go out and generate additional data describing each of these samples, we have to look at proteins, and we have to look at metabolites. On the other hand, it also makes things easier in some sense because we’ll have a lot more context with which to address the question.”
To do that well, they will need exceptional analytical tools and methods. “When these data really start to arrive in bulk, we’ll need new, efficient, informative ways of tacking together information that isn’t just about one bug or about one bug and a host, but about a whole community of bugs over time,” Huttenhower says. They are working with colleagues to develop computational models themselves, but will be on the lookout for other tools that might be helpful too. He adds, “We’ll use anything we can get our hands on.”
In Sickness and in Health
Huttenhower and other microbiome researchers are the first to say that the field is young and only beginning to appreciate the contributions of the approximately 2.5 lb of bacteria and other microbes that share our bodies with us.
“So here we are in 2014. We’ve learned a huge amount about the composition of the microbiome. We have a pretty good idea about who’s there, but we’re just getting started on what they’re doing,” Proctor says, noting that many projects are already providing glimpses of the potential for clinical applications. For instance, she pointed to the work of a Cleveland Clinic research group that reported that compounds produced when the gut microbiome metabolizes red meat may actually be better markers for cardiovascular disease than cholesterol levels [5]. She believes that work like this will give the microbiome the attention it deserves.
White agrees. “I believe the microbiome is creating a unique opportunity for us to think about the factors that you can adjust to remain healthier or to become healthier,” he says. “Just as we were blind to the importance of low levels of iodine in the diet not all that long ago, today we are blind to the importance of a healthy microbiome. We’re learning that by taking incredibly simple steps, we can probably improve the health of our population in a big way.”
White adds, “I can imagine that in five to ten years’ time, we will be able to elect to look at our gut microbiome and just order off the shelf a cocktail of healthy bacteria that we can then ingest to use for such things as losing weight or reducing the risk of getting asthma, or possibly to protect us from different types of cancer or from immunological diseases.”
As understanding expands and that knowledge is applied to the clinic, the old view of the microbiome as just a bunch of “bad bugs” is starting to fade away. In its place is a new perception of the microbiome as sometimes participating in disease, but also vital to health. “The biomedical community is realizing that it’s missed a major part of the story,” Proctor asserts, noting that microbes are only now starting to get their due. “Yes, some microbes do have a role in disease and we need to understand that role, but what’s probably even more mind-boggling is that we also need microbes. They are an essential part of our health and well-being.”
References
- K. Aagaard, M. Jun, K. M. Antony, G. Radhika, J. Petrosino, and J. Versalovic, “The placenta harbors a unique microbiome,” Sci. Transl. Med., vol. 6, no. 237, pp. 237–265, May 2014.
- The Human Microbiome Project Consortium, “A framework for human microbiome research,” Nature, vol. 486, no. 7402, pp. 215–221, June 2012.
- NIH Human Microbiome Project Tools & Technology. [Online].
- NIH Human Microbiome Project Funded Research. (2014, May 6). [Online].
- R. A. Koeth, Z. Wang, B. S. Levison, J. A. Buffa, E. Org, B. T. Sheehy, E. B. Britt, X. Fu, Y. Wu, L. Li, J. D. Smith, J. A. DiDonato, J. Chen, H. Li, G. D. Wu, J. D. Lewis, M. Warrier, J. M. Brown, R. M. Krauss, W. H. W. Tang, F. D. Bushman, A. J. Lusis, and S. L. Hazen, “Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis,” Nat. Med., vol. 19, no. 5, pp. 576–585, May 2013.