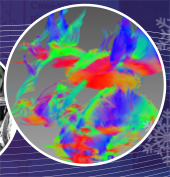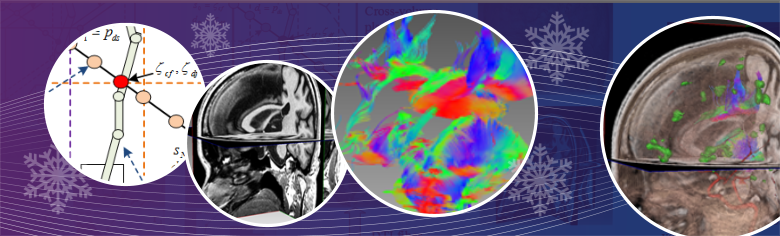
Brain lesions are usually located adjacent to critical spinal structures, so it will be a challenging task for neurosurgeons to precisely plan a surgical procedure without damaging healthy tissues and nerves. The advancement of medical imaging technologies produces a large amount of neurological data, which are capable of showing a wide variety of brain properties. Advanced algorithms of data computing and visualization are critically helpful in efficiently utilizing the acquired data for disease diagnosis and treatment planning. In this paper, we describe new algorithms and a software framework for multiple volume of interest specified diffusion tensor imaging (DTI) fiber dynamic visualization. We first set multiple flexible cube-based VOIs and then developed new algorithms for DTI fiber visualization. In our system, the DTI fibers were tracked in real time and were dynamically specified by multiple VOIs for interactively analyzing fiber connectivity. Next, the multi-function neurological images were volume rendered on graphics hardware and fused with the dynamically displayed fiber tracks in multiple synchronized rendering windows. In the following step, 3D data visualization and fusion were accomplished with GPU-based raycasting and a novel voxel classification algorithm, which offered neurosurgeons with interactivity and flexibility to see the targets from viewpoints not physically possible and free them from mentally reconstructing 3D images from 2D slices. Finally, a depth texture indexing algorithm was used to detect DTI fiber tracts in GPUs, which makes fibers to be displayed and interactively manipulated with brain data acquired from functional magnetic resonance imaging (fMRI), T1- and T2-weighted anatomic imaging, and angiographic imaging.
The developed software platform is built on an object-oriented structure, which is transparent and extensible. It provides a comprehensive human-computer interface for data exploration and information extraction. The GPU-accelerated high-performance computing kernels have been implemented to enable our software to dynamically visualize neurological data. The developed techniques will be useful in computer-aided neurological disease diagnosis, brain structure exploration, and general cognitive neuroscience.