Artificial intelligence is powering early diagnosis of breast cancer risk, streamlining medical care in hospitals, and helping to identify rare diseases
Technologies to provide early predictions of breast cancer risk, to identify which hospital patients actually should have their vital signs monitored overnight and which should be left to their restorative sleep, and to swiftly identify rare infant diseases are all joining the ranks of approaches that are powered by artificial intelligence (AI).
By making use of the vast amounts of digitized data that are now collected, tools like these provide a clear window into the ever-broadening avenues through which AI will touch health care. In the first half of 2021 alone, for instance, the U.S. Food and Drug Administration (FDA) authorized dozens of AI-enabled devices to allow for early diagnoses of disease, streamline medical care, and personalize treatments [1].
The three described here are a quick glimpse into the variety of projects under way, the challenges in their development, and their progress from idea to clinically used product.
Predicting breast cancer
One of the best ways to fight breast cancer is to catch it early, and a new AI model called Mirai is designed to assess a woman’s risk of developing the disease, and do so as much as five years before a mammogram would identify it.
In late November 2021, a validation study performed at seven hospitals spanning seven countries and four continents, showed that this deep-learning model was able to predict the one- to five-year risk of breast cancer from mammograms with an overall 44% accuracy [2]. “The model maintained consistent performance across all the different medical centers,” said Adam Yala, the first author of the study article [2], and a doctoral student in the research group of Regina Barzilay, Ph.D., professor of electrical engineering and computer science at the Massachusetts Institute of Technology (MIT) (Figure 1). “The next stage is prospective trials and that is what we’re working on launching now with all of our clinical partners,” Yala added.

To build Mirai, the researchers trained it on more than 200,000 standard, digital, two-dimensional (2-D) mammograms gathered from Massachusetts General Hospital (MGH) patients who eventually were diagnosed with breast cancer. It involved more than simply uploading the images, Yala said. The MGH mammograms were collected from multiple mammogram machines, and although the machines may each be accurate, they might not be calibrated the same. That, he said, could throw off the risk model. “So rather than just training the model to predict risk, we trained the model to figure out which machine the mammogram came from, and by doing so, we could fully eliminate the device bias while still maintaining the model’s accuracy in risk prediction.”
Beyond that, the researchers looked at risk-factor data gathered from patients. MGH collects very detailed information on risk factors from patients, but other health care facilities may not always have that same capacity, Yala explained. To navigate that disparity, the researchers used the MGH patient data to train the model, but then developed the model so it could make predictions from the mammograms directly. “That’s why this model can actually be used at all these different hospitals,” he noted.
Finally, the research group considered the training mammograms at different time points. The group first predicted the one-year risk, which was clearest because the number of mammograms becomes progressively fewer in subsequent years [3]. They then used the one-year risk to help predict the two-year risk, and did the same with the two-year data to predict the three-year risk, and so on up to the five-year point. “If you look at a five-year risk model, you’re losing a lot of the data that build up in five years, so this was a holistic way to model all the available data at the same time, and leverage the relationship between different time points,” Yala said.
With the retrospective validation of the model completed, the research group is now preparing to launch trials with a range of clinical partners, he said. “Our clinical partners have their own visions of how this could help the clinical workflow, but one of our overarching objectives is to update current clinical guidelines about which patients should get high-risk screening, such as magnetic resonance imaging (MRI).” Yala added, “We’re still in the ‘active design stage’ because the different clinical trials that will help build evidence have their own local constraints (including insurance coverage of MRIs), but that is our ultimate goal.”
Check or sleep
Let Sleeping Patients Lie is the appropriate name of AI technology that delves into the common practice of taking the vital signs—checking blood pressure, temperature, respiration, and pulse rate—every three top 5 hours, even if it means waking patients in the middle of the night.
Not surprisingly, patients do not like the practice, said Theodoros Zanos, Ph.D., head of the Neural and Data Science Laboratory, Feinstein Institutes for Medical Research, Northwell Heath, Manhasset, NY, USA (Figure 2). “We looked back at patient feedback at Northwell Health, which is the largest health system in New York, and found that a top patient complaint is that they do not sleep well during their stay in the hospital, and that is partly because all patients, including those who are stable, are woken up overnight to take their vitals.” Patient feedback at other hospitals shows the same thing.

“This disruption of sleep not only affects the patient experience, but many studies also show that sleep is actually a critical piece of helping patients get better, especially in the hospital,” he reported, noting that Northwell clinicians and nurses agreed. “In addition, poor sleep is related to other problems, such as an increase in the chance of patient falls particularly in the older population, or even delirium in certain patients. So overall, it’s not a good idea to wake up patients unless you really have to.”
The research groups of Zanos and collaborator Jamie Hirsch, M.D., a physician at Northwell’s Great Neck Medical Center, set out to develop an algorithm that could determine which patients could go without the overnight vitals check. They trained and evaluated the algorithm on electronic-health-record data collected from some 2.3 million patients who had passed through Northwell’s hospitals between 2012–2019, Zanos explained [4]. “From that, we were able to develop an algorithm that uses the age and body mass index, and tracks the trends of each patient’s vitals as they develop over the previous one or two days, and then at the specific time point, say at 10 p.m., it produces a report for the nurses with predictions of which patients are safe to be left sleeping overnight” (Figure 3).
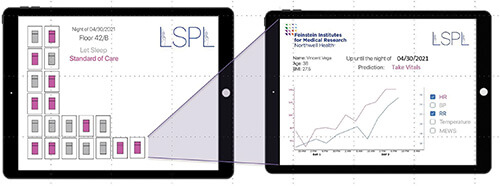
In developing Let Sleeping Patients Lie, the researchers set a lofty goal so it wouldn’t inappropriately place a patient in the do-not-wake column. “Through discussions again with several physicians, we arrived at a figure that we can only miss two out of 10,000 nights, which is pretty strict for all intents and purposes, and the algorithm achieves that,” Zanos said, noting that they just completed more than a year of extensive background validation of the algorithm at Northwell’s hospitals. They have also just started live testing in a pilot study at Northwell’s Huntington Hospital, Huntington, NY, USA, and hope to add more hospital pilot studies to learn how nurses are using it; and whether it changes hospital workflow by freeing up nurses to focus their overnight attention on those patients who need it, affects patient outcomes or lengths of stay, or boosts general patient satisfaction.
Future work also includes incorporating data from continuous monitoring devices into the algorithm as a way of expanding it so it does not only identify patients who are doing well at night, but also those who have a risk of deteriorating throughout the entire day.
Zanos and other researchers also hope to expand the approach to other predictive models, and hospitals are working on using a version to predict respiratory failure among COVID-19 patients within 48 hours [5].
Of AI’s overall role in health care, he remarked, “As long as it is done in the right way methodologically and with lots of data, which is something we have as part of a very large health system, we really think that this combination of AI and medicine is very promising in assisting medical decision-making.”
Spotting rare disease
AI also presents an opportunity to make a big contribution to the diagnosis of rare diseases, which, according to the U.S. definition, number around 7000 [6]. To that end, researchers at Fabric Genomics, Oakland, CA, USA, have designed an approach that allows for the swift diagnosis of rare diseases, particularly among sick infants in the neonatal intensive care unit (NICU).
The algorithm at the heart of the technology, called Fabric GEM, essentially mimics the months-long work that a pediatrician and clinical geneticist have previously had to do to come up with a diagnosis, but does it automatically and much faster, according to computational biologist and geneticist Martin Reese, Ph.D., the company’s co-founder, president, and chief executive officer (Figure 4). Currently, using GEM, doctors are able to return a diagnosis in two to three days, and in some NICU cases, can drop the time to just 4 hours—sometimes even less, he said. “And as you can imagine, that’s really what’s necessary to help the kids in the NICU who need a diagnosis within a day or two.”

He described GEM as “a classic application of a massive AI system” that stands out because it couples the patient’s genotype with phenotype—the symptoms and clinical data—and by doing so, homes in only on relevant mutations. “Everyone has mutations that are silent, or that may be important for you later in life or for your offspring, but not for the immediate diagnosis, so that linking of the phenotype to the genomic diagnostic process was the absolute breakthrough for us,” Reese said.
To do that, the researchers needed previously gathered data about both the genotypes and phenotypes associated with rare diseases, but because these diseases occur so infrequently, that information is scattered in databases around the world. Tracking down the data was not the only issue. Information is often collected using methods that are not directly comparable, and much of it is stored in clinician notes that demand analysis with natural language processing software, he explained. In addition, Reese said, “The biological and clinical data are academic and very error-prone, so we have a whole quality-control team at Fabric Genomics that double checks every new piece of data for consistency and to make sure it’s actually real.”
A retrospective study published late last year showed that GEM was more than 90% successful in identifying a short list of genetic variants, usually just one or two, that were associated with an infant’s rare genetic disease [7]. The study used whole-genome and whole-exome data from 119 patients at Rady Children’s Institute for Genomic Medicine, San Diego, CA, USA, and 60 additional patients at several other clinical centers, including Boston Children’s Hospital, Hudson Alpha Christian-Albrechts University, Kiel, Germany, and Tartu University Hospital, Tartu, Estonia.
With that vote of confidence, Fabric Genomics now hopes to make GEM even more helpful by not only providing doctors with candidate diagnoses, but also linking them to treatment plans that have been and are being developed by the medical community. “There are consortia, including at the NIH, that are putting together treatment plans for these illnesses, so we will link to what basically will be a new textbook for 7000 rare genetic diseases,” Reese said.
“This is very exciting,” he remarked. “There are so many single-gene-named organizations out there already, and parents are continuing to form groups and push for new treatments and funding, so I think the entire rare-disease community will benefit from this approach in a massive way.”
References
- U.S. Food and Drug Administration. (Sep. 22, 2021). Artificial Intelligence and Machine Learning (AI/ML)-Enabled Medical Devices. Accessed: Dec. 20, 2021. [Online]. Available: https://www.fda.gov/medical-devices/software-medical-device-samd/artificial-intelligence-and-machine-learning-aiml-enabled-medical-devices
- A. Yala et al., “Multi-institutional validation of a mammography-based breast cancer risk model,” J. Clin. Oncol., Nov. 2021. Accessed: Dec. 27, 2021. [Online]. Available: https://ascopubs.org/doi/full/10.1200/JCO.21.01337, doi: 10.1200/JCO.21.01337.
- A. Yala et al., “Toward robust mammography-based models for breast cancer risk,” Sci. Transl. Med., vol. 13, no. 578, Jan. 2021, Art. no. eaba4373.
- V. Tóth et al. “Let sleeping patients lie, avoiding unnecessary overnight vitals monitoring using a clinically based deep-learning model,” NPJ Digit. Med., vol. 3, p. 149, Nov. 2020. Accessed: Dec. 31, 2021, doi: 10.1038/s41746-020-00355-7.
- S. Bolourani et al., “A machine learning prediction model of respiratory failure within 48 hours of patient admission for COVID-19: Model development and validation,” J. Med. Internet Res., vol. 23, no. 2, p. e24246, Feb. 2021. Accessed: Dec. 31, 2021. [Online]. Available: https://www.jmir.org/2021/2/e24246/
- U.S. National Institutes of Health. FAQs About Rare Diseases. Accessed: Dec. 29, 2021. [Online]. Available: https://rarediseases.info.nih.gov/diseases/pages/31/faqs-about-rare-diseases
- F. M. De La Vega et al., “Artificial intelligence enables comprehensive genome interpretation and nomination of candidate diagnoses for rare genetic diseases,” Genome Med., vol. 13, p. 153, Oct. 2021. Accessed: Dec. 31, 2021, doi: 10.1186/s13073-021-00965-0.






